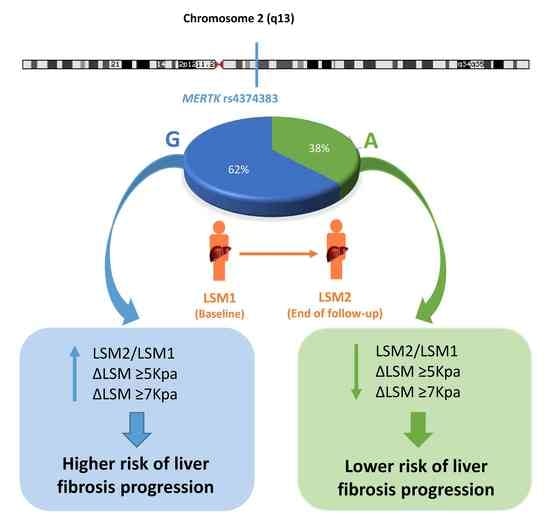
The Myeloid-Epithelial-Reproductive Tyrosine Kinase (MERTK) rs4374383 Polymorphism Predicts Progression of Liver Fibrosis in Hepatitis C Virus-Infected Patients: A Longitudinal Study
Abstract
:1. Introduction
2. Patients and Methods
2.1. Study Population
2.2. Clinical Data
2.3. HCV Assays
2.4. Genotyping of MERTK SNP
2.5. Liver Stiffness Measurement
2.6. Outcome Variable
2.7. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Study Population
3.2. Characteristics of the MERTK rs4374383 SNP
3.3. MERTK rs4374383 SNP and Liver Fibrosis Progression
4. Discussion
5. Limitations of Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Westbrook, R.H.; Dusheiko, G. Natural history of hepatitis C. J. Hepatol. 2014, 61, S58–S68. [Google Scholar] [CrossRef] [PubMed]
- European Association for Study of Liver. EASL recommendations on treatment of hepatitis C 2015. J. Hepatol. 2015, 63, 199–236. [Google Scholar] [CrossRef] [PubMed]
- Tsochatzis, E.A.; Bosch, J.; Burroughs, A.K. Liver cirrhosis. Lancet 2014, 383, 1749–1761. [Google Scholar] [CrossRef]
- Resino, S.; Sanchez-Conde, M.; Berenguer, J. Coinfection by human immunodeficiency virus and hepatitis C virus: Noninvasive assessment and staging of fibrosis. Curr. Opin. Infect. Dis. 2012, 25, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Castera, L. Noninvasive assessment of liver fibrosis. Dig. Dis. 2015, 33, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Castera, L.; Vergniol, J.; Foucher, J.; Le Bail, B.; Chanteloup, E.; Haaser, M.; Darriet, M.; Couzigou, P.; De Ledinghen, V. Prospective comparison of transient elastography, fibrotest, apri, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology 2005, 128, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Rueger, S.; Bochud, P.Y.; Dufour, J.F.; Mullhaupt, B.; Semela, D.; Heim, M.H.; Moradpour, D.; Cerny, A.; Malinverni, R.; Booth, D.R.; et al. Impact of common risk factors of fibrosis progression in chronic hepatitis C. Gut 2015, 64, 1605–1615. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.H.; Bochud, P.Y.; George, J. Host-hepatitis C viral interactions: The role of genetics. J. Hepatol. 2016, 65, S22–S32. [Google Scholar] [CrossRef] [PubMed]
- Van der Meer, J.H.; van der Poll, T.; van ’t Veer, C. Tam receptors, gas6, and protein S: Roles in inflammation and hemostasis. Blood 2014, 123, 2460–2469. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.K.; Wilhelm, A.; Antoniades, C.G. Tam receptor tyrosine kinase function and the immunopathology of liver disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G899–G905. [Google Scholar] [CrossRef] [PubMed]
- De Minicis, S.; Seki, E.; Uchinami, H.; Kluwe, J.; Zhang, Y.; Brenner, D.A.; Schwabe, R.F. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology 2007, 132, 1937–1946. [Google Scholar] [CrossRef] [PubMed]
- Kupcinskas, J.; Valantiene, I.; Varkalaite, G.; Steponaitiene, R.; Skieceviciene, J.; Sumskiene, J.; Petrenkiene, V.; Kondrackiene, J.; Kiudelis, G.; Lammert, F.; et al. PNPLA3 and RNF7 gene variants are associated with the risk of developing liver fibrosis and cirrhosis in an eastern european population. J. Gastrointestin. Liver Dis. 2017, 26, 37–43. [Google Scholar] [PubMed]
- Petta, S.; Valenti, L.; Marra, F.; Grimaudo, S.; Tripodo, C.; Bugianesi, E.; Camma, C.; Cappon, A.; Di Marco, V.; Di Maira, G.; et al. MERTK rs4374383 polymorphism affects the severity of fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2016, 64, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Patin, E.; Kutalik, Z.; Guergnon, J.; Bibert, S.; Nalpas, B.; Jouanguy, E.; Munteanu, M.; Bousquet, L.; Argiro, L.; Halfon, P.; et al. Genome-wide association study identifies variants associated with progression of liver fibrosis from HCV infection. Gastroenterology 2012, 143, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, R.S.; Dasarathy, S.; McCullough, A.J. Alcoholic liver disease. Hepatology 2010, 51, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Calvaruso, V.; Craxì, A. 2011 european association of the study of the liver hepatitis C virus clinical practice guidelines. Liver Int. 2012, 32, 2–8. [Google Scholar] [CrossRef] [PubMed]
- European Association for Study of Liver. EASL clinical practice guidelines: Management of hepatitis C virus infection. J. Hepatol. 2014, 60, 392–420. [Google Scholar] [CrossRef] [PubMed]
- Sandrin, L.; Fourquet, B.; Hasquenoph, J.M.; Yon, S.; Fournier, C.; Mal, F.; Christidis, C.; Ziol, M.; Poulet, B.; Kazemi, F.; et al. Transient elastography: A new noninvasive method for assessment of hepatic fibrosis. Ultrasound Med. Biol. 2003, 29, 1705–1713. [Google Scholar] [CrossRef] [PubMed]
- Albert, P.S. Longitudinal data analysis (repeated measures) in clinical trials. Stat. Med. 1999, 18, 1707–1732. [Google Scholar] [CrossRef]
- Senn, S.; Stevens, L.; Chaturvedi, N. Repeated measures in clinical trials: Simple strategies for analysis using summary measures. Stat. Med. 2000, 19, 861–877. [Google Scholar] [CrossRef]
- Miyaaki, H.; Ichikawa, T.; Taura, N.; Miuma, S.; Honda, T.; Shibata, H.; Soyama, A.; Hidaka, M.; Takatsuki, M.; Eguchi, S.; et al. Impact of donor and recipient single nucleotide polymorphisms in living liver donor transplantation for hepatitis C. Transplant. Proc. 2015, 47, 2916–2919. [Google Scholar] [CrossRef] [PubMed]
- Musso, G.; Cassader, M.; De Michieli, F.; Paschetta, E.; Pinach, S.; Saba, F.; Bongiovanni, D.; Framarin, L.; Berrutti, M.; Leone, N.; et al. MERTK rs4374383 variant predicts incident nonalcoholic fatty liver disease and diabetes: Role of mononuclear cell activation and adipokine response to dietary fat. Hum. Mol. Genet. 2017, 26, 1747–1758. [Google Scholar] [CrossRef] [PubMed]
- Pastore, M.; Maira, G.D.; Galigiuri, A.; Petta, S.; Marra, F. Role of the protein tyrosine kinase mer (MerTK) in the cross-talk between macrophages and hepatic stellate cells. J. Hepatol. 2018, 68, S407–S408. [Google Scholar] [CrossRef]
- Dransfield, I.; Zagorska, A.; Lew, E.D.; Michail, K.; Lemke, G. Mer receptor tyrosine kinase mediates both tethering and phagocytosis of apoptotic cells. Cell Death Dis. 2015, 6, e1646. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Keating, A.K.; Joung, D.; Sather, S.; Kim, G.K.; Sawczyn, K.K.; Brandao, L.; Henson, P.M.; Graham, D.K. Mer tyrosine kinase (MerTK) promotes macrophage survival following exposure to oxidative stress. J. Leukoc. Biol. 2009, 86, 73–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli, M.; Pan, G.; Nord, H.; Wallen Arzt, E.; Wallerman, O.; Wadelius, C. Genetic prevention of hepatitis C virus-induced liver fibrosis by allele-specific downregulation of MERTK. Hepatol. Res. 2017, 47, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Tamura, T.; Ishihara, M.; Lamphier, M.S.; Tanaka, N.; Oishi, I.; Aizawa, S.; Matsuyama, T.; Mak, T.W.; Taki, S.; Taniguchi, T. An irf-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated t lymphocytes. Nature 1995, 376, 596–599. [Google Scholar] [CrossRef] [PubMed]
- Carmona, I.; Cordero, P.; Ampuero, J.; Rojas, A.; Romero-Gomez, M. Role of assessing liver fibrosis in management of chronic hepatitis C virus infection. Clin. Microbiol. Infect. 2016, 22, 839–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aleman, S.; Rahbin, N.; Weiland, O.; Davidsdottir, L.; Hedenstierna, M.; Rose, N.; Verbaan, H.; Stal, P.; Carlsson, T.; Norrgren, H.; et al. A risk for hepatocellular carcinoma persists long-term after sustained virologic response in patients with hepatitis C-associated liver cirrhosis. Clin. Infect. Dis. 2013, 57, 230–236. [Google Scholar] [CrossRef] [PubMed]


| Characteristic | All Patients | MERTK rs4374383 SNP | ||
|---|---|---|---|---|
| AA | AG | GG | ||
| No. | 208 | 25 | 109 | 74 |
| Male | 112 (53.8%) | 15 (60%) | 59 (54.1%) | 38 (51.4%) |
| Age (years) | 47.1(41.5; 57.6) | 42.3 (38.2; 49.1) | 47.6 (42.2; 59.5) | 49.1 (42.7; 58.7) |
| Time of HCV infection (years) | 8.2 (3.2; 13.2) | 7.2 (3.9; 16.7) | 9.8 (3.3; 13.3) | 6.4 (2.7; 12.1) |
| High alcohol intake | 28 (13.5%) | 6 (24%) | 13 (11.9%) | 9 (12.1%) |
| Prior injection drug use | 21 (10.1%) | 6 (24%) | 8 (7.3%) | 7 (9.5%) |
| HCV genotype (n = 204) | ||||
| 1 | 174 (85.3%) | 17 (68%) | 91 (86.7%) | 66 (89.2%) |
| 3 | 14 (6.9%) | 4 (16%) | 7 (6.5%) | 3 (4.1%) |
| 4 | 15 (7.4%) | 4 (16%) | 6 (5.7%) | 5 (6.8%) |
| 5 | 1 (0.5%) | 0 (0%) | 1 (1%) | 0 (0%) |
| Prior failed peg-IFN-α/RBV therapy | 47 (22.6%) | 4 (16%) | 30 (27.5%) | 13 (17.6%) |
| Baseline LSM (kPa) | 6.1 (5.2; 7.7) | 6.4 (4.6; 7.4) | 6.3 (5.1; 8.0) | 6 (5.3; 6.8) |
| F0–F1 (<7.1 kPa) | 149 (71.6%) | 17 (68%) | 72 (66.1%) | 60 (81.1%) |
| F2 (7.1–9.4 kPa) | 38 (18.3%) | 6 (24%) | 25 (22.9%) | 7 (9.5%) |
| F3 (9.5–12.4 kPa) | 21 (10.1%) | 2 (8%) | 12 (11%) | 7 (9.5%) |
| Follow-up time (months) | 46.6 (28.7; 61.5) | 48.6 (30.1; 64.1) | 47.9 (28.9; 60.6) | 45.2 (25.5; 61.5) |
| Final LSM (kPa) | 6.8 (5.5; 9.4) | 6.3 (5.1; 8.7) | 7.4 (5.9; 10.1) | 6.8 (5.4; 8.9) |
| F0–F1 (<7.1 kPa) | 110 (52.9%) | 17 (68%) | 53 (48.6%) | 40 (54.1%) |
| F2 (7.1–9.4 kPa) | 47 (22.6%) | 3 (12%) | 28 (25.7%) | 16 (21.6%) |
| F3 (9.5–12.4 kPa) | 25 (12%) | 5 (20%) | 13 (11.9%) | 7 (9.5%) |
| F4 (≥12.5 kPa) | 26 (12.5%) | 0 (0%) | 15 (13.8%) | 11 (14.9%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Sousa, M.Á.; Gómez-Moreno, A.Z.; Pineda-Tenor, D.; Brochado-Kith, O.; Sánchez-Ruano, J.J.; Artaza-Varasa, T.; Gómez-Sanz, A.; Fernández-Rodríguez, A.; Resino, S. The Myeloid-Epithelial-Reproductive Tyrosine Kinase (MERTK) rs4374383 Polymorphism Predicts Progression of Liver Fibrosis in Hepatitis C Virus-Infected Patients: A Longitudinal Study. J. Clin. Med. 2018, 7, 473. https://doi.org/10.3390/jcm7120473
Jiménez-Sousa MÁ, Gómez-Moreno AZ, Pineda-Tenor D, Brochado-Kith O, Sánchez-Ruano JJ, Artaza-Varasa T, Gómez-Sanz A, Fernández-Rodríguez A, Resino S. The Myeloid-Epithelial-Reproductive Tyrosine Kinase (MERTK) rs4374383 Polymorphism Predicts Progression of Liver Fibrosis in Hepatitis C Virus-Infected Patients: A Longitudinal Study. Journal of Clinical Medicine. 2018; 7(12):473. https://doi.org/10.3390/jcm7120473
Chicago/Turabian StyleJiménez-Sousa, María Ángeles, Ana Zaida Gómez-Moreno, Daniel Pineda-Tenor, Oscar Brochado-Kith, Juan José Sánchez-Ruano, Tomas Artaza-Varasa, Alicia Gómez-Sanz, Amanda Fernández-Rodríguez, and Salvador Resino. 2018. "The Myeloid-Epithelial-Reproductive Tyrosine Kinase (MERTK) rs4374383 Polymorphism Predicts Progression of Liver Fibrosis in Hepatitis C Virus-Infected Patients: A Longitudinal Study" Journal of Clinical Medicine 7, no. 12: 473. https://doi.org/10.3390/jcm7120473