- 1Unit of Human Parasite Molecular and Cell Biology, Key Laboratory of Molecular Virology and Immunology, Institut Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Clinical Laboratory Medicine, Changzhi People’s Hospital, Changzhi, China
- 4Department of Parasitology, School of Basic Medical Science, Wenzhou Medical University, Wenzhou, China
- 5Institut de Médecine Régénératrice et de Biothérapie, INSERM U1183, CHU Montpellier, Montpellier, France
- 6ShanghaiTech University, Shanghai, China
Plasmodium falciparum erythrocyte membrane protein 1, encoded by var gene, is an immunodominant antigen mediating immune evasion in humans. At a given time, only a single var gene is commonly expressed in one parasite. However, the regulation mechanism of var transcription remains largely unknown. In this study, we identified the antisense long non-coding RNA (aslncRNA) derived from var intron as an activation factor for the corresponding var gene. The exogenous artificial var aslncRNA transcribed by T7 RNA polymerase from episome can specifically activate the homologous var gene, and the exogenous aslncRNA activates transcription of both var mRNA and endogenous aslncRNA in a manner independent of the conserved intron sequence within the var gene family. Interestingly, the newly activated var gene and the previously dominant var gene then could be co-expressed in the same parasite nuclei, which suggests that the aslncRNA-mediated var gene activation could escape from the control of mutually exclusively expression of the var gene family. Together, our work shows that var aslncRNA is the activator responsible for var gene transcriptional regulation.
Introduction
Plasmodium falciparum, one pathogen of malaria, caused more than 200 million infections and 445,000 deaths in 2016 (WHO, 2017). It parasitizes and reproduces in erythrocytes of infected individuals. On the surface of infected erythrocytes, multiple proteins can be expressed and presented by the malaria parasite, such as erythrocyte membrane protein 1 (PfEMP1), Rifin, Stevor (Dzikowski et al., 2006). PfEMP1, as an immunodominant antigen of P. falciparum (Leech et al., 1984), encoded by var gene, possesses the characteristic of antigen polymorphism. This protein is attributed with parasite escape from the host immune system and allows infected erythrocytes to adhere onto uninfected erythrocytes, a process related to disease severity (Rowe et al., 1995).
In P. falciparum 3D7, there are about 60 var gene members with several similar gene features, including polymorphic exonI, conserved exonII and a bi-directional promoter-activity in the intron (Calderwood et al., 2003). It is reported that only one var gene is exclusively expressed at ring stage of a single parasite cell, while other var genes are silent, referred to mutually exclusive gene expression (MEE) (Scherf et al., 1998). This phenomenon has also been found in antigen families of other parasite species (Pays et al., 2004; Prucca et al., 2008). Two main mechanisms are widely accepted for MEE regulation of multiple gene families in many other species: DNA rearrangement and epigenetic modification (Deitsch et al., 2009). However, it has been demonstrated that, in P. falciparum, DNA rearrangement and RNA interference could not be the main mechanism of var gene expression regulation (Scherf et al., 1998; Baum et al., 2009). Many other factors have been found to contribute to the regulation of var gene transcription, such as cis-elements (Deitsch et al., 2001; Calderwood et al., 2003), trans-factors (Voss et al., 2003; Fraschka et al., 2016), epigenetic markers (Lopez-Rubio et al., 2007; Volz et al., 2012; Jiang et al., 2013), and higher order chromatin structure (Duraisingh et al., 2005; Ralph et al., 2005b). Nevertheless, the whole MEE mechanism of var gene still remains unclear.
Recently, var gene antisense long non-coding RNAs (aslncRNAs) have emerged as new regulating candidates for var gene transcription. The var aslncRNAs are originated from var intron and extended to exonI. They are retained in the nucleus and are regarded as potential regulators (Epp et al., 2009). Previous studies indicate that the transcription of a particular aslncRNA correlated with the activation of the corresponding var gene (Jiang et al., 2013; Avraham et al., 2015). And other evidences suggest that aslncRNA is involved in the activation of var gene promoter (Avraham et al., 2015). However, Ralph and collaborators state that the silencing or activation of var gene is not overall correlated with these antisense sterile transcripts (Ralph et al., 2005a). In addition, the intron deletion of the var2csa, a conserved member of var family, could promote its transcriptional level, which also implies that the aslncRNA is not essential for var gene activation (Bryant et al., 2017). Therefore, it is still a controversial issue whether the var aslncRNA is involved in var transcription regulation.
To identify the role of aslncRNA in the var gene activation, artificial var aslncRNAs in this study were generated using T7 RNA polymerase in P. falciparum, and the var expression patterns were analyzed. We found that the artificial aslncRNA could specifically induce the corresponding var gene transcription, and the exonI region of this sterile transcript was capable of var specific activation. Additionally, transcription of the artificial var aslncRNA also induced the aslncRNA promoter activity of the corresponding var intron. RNA-FISH of var mRNA indicated that the previously dominant and newly induced var gene mRNAs could exist in one parasite. These findings demonstrated that the var aslncRNA exerted activatory function on var gene transcription and provided a theoretical basis for further researches on var MEE regulation.
Materials and Methods
Parasites in vitro Culture and Transfection
Plasmodium falciparum 3D7 strain C8, G4, and P. falciparum NF54 strain A3 were cultured as described previously (Cranmer et al., 1997). The plasmid was transfected into the parasites by electrotransformation (Gene Pulser Xcell, BIO-RAD) as mentioned before (Rug and Maier, 2012), and 2 μg/ml blasticidin S (BSD, Invitrogen) was used for transformants selection.
Mapping 5′ End of var aslncRNA
The 5′ end RACE (rapid amplification of cDNA ends) of the PF3D7_0617400 aslncRNA was performed with SMART RACE cDNA kit (TAKARA) according to the manufacturer’s instructions. P. falciparum NF54 strain A3 was harvested at ring stage, and the total RNA was extracted with TRIzol (Invitrogen). Five microgram total RNA of A3 strain was prepared for 5′ end determination of PF3D7_0617400 aslncRNA. The primer for 5′ end identification is listed in Supplementary Table S1.
Reverse Transcription Quantitative PCR (RT-qPCR)
One microgram of the extracted total RNA of P. falciparum was reverse transcribed using FastQuant RT Kit (Tiangen) according to its standard manuals. The RT-qPCR was performed as described (Salanti et al., 2003), and serine-tRNA ligase (PF3D7_0717700) was used as an internal control. All the RT-qPCR primer pairs for mRNA or lncRNA detection are listed in Supplementary Table S1.
Construction of var aslncRNA Expression Vectors
To construct pCC4-NLS-T7RNP, the coding sequence of T7 RNA polymerase was amplified from Escherichia coli BL21(DE3) genome and inserted between XhoI and SmaI sites of pCC4 (Maier et al., 2006). The nucleus localization signal sequence of yeast Gal4p (1–222 bp) from pGKBT7 (Clonetech) and Flag-tag were fused on the N-terminal of T7 RNA polymerase. This fusion protein was named NLS-T7RNP. T7 promoter and terminator were amplified by overlapping PCR and cloned into pCC4-NLS-T7RNP at the AvrII site, which was named “pT7SE.” According to the description of PF3D7_0617400 aslncRNA in (Epp et al., 2009), different artificial PF3D7_0617400 aslncRNA templates, almost covering natural aslncRNA, were, respectively, inserted in SmaI restriction site downstream of T7 promoter (Figure 1A). In these plasmids, the bsd gene can be expressed in the P. falciparum and confer resistance of BSD. Plasmids were constructed by In-Fusion technology (Vazyme). All primers used for plasmids construction are listed in Supplementary Table S1. The transcriptional activity of NLS-T7RNP was confirmed by additional experiments in E. coli (Supplementary Figure S1).
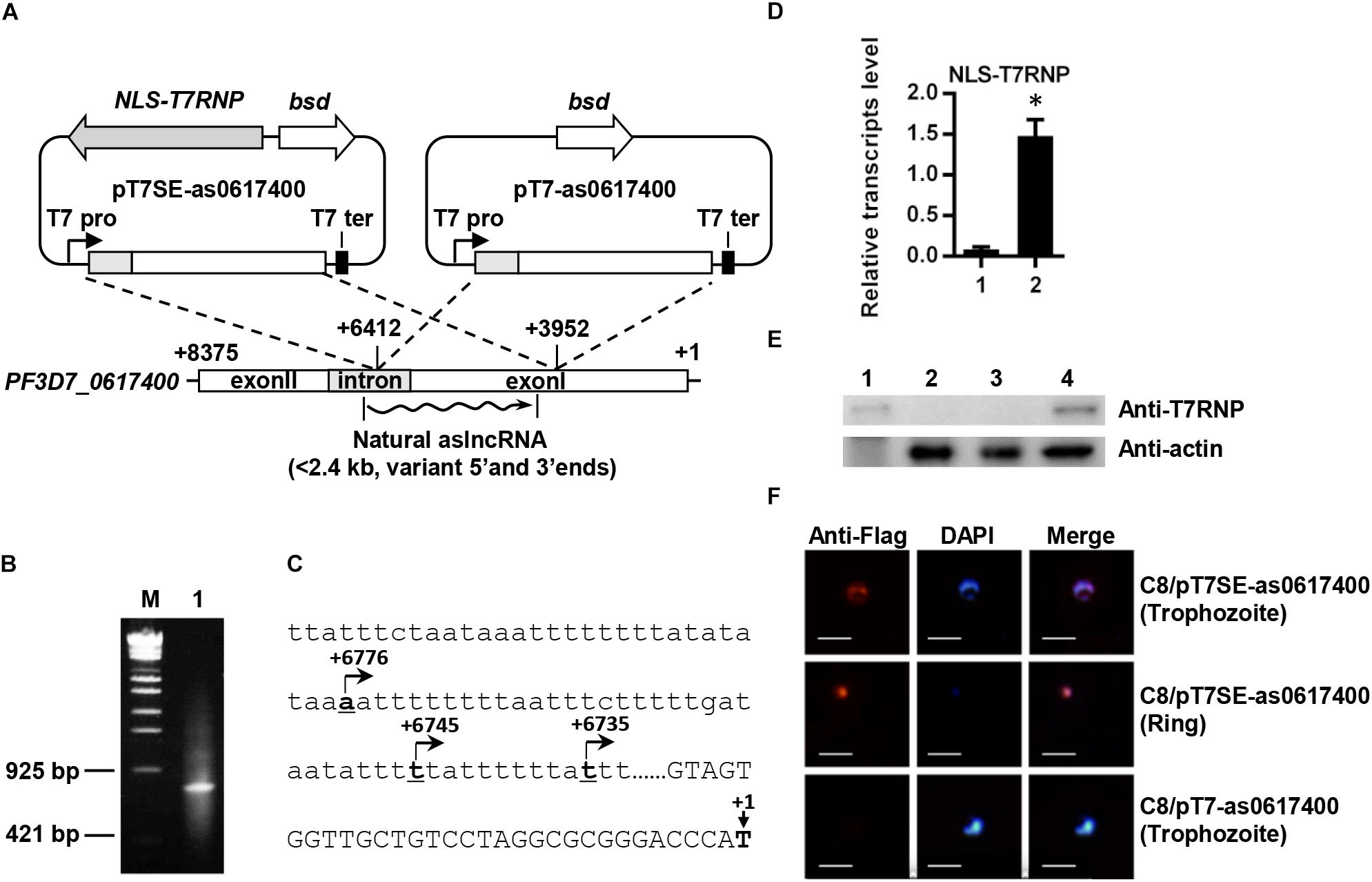
Figure 1. Schematic diagram of plasmids for expressing the var aslncRNA and 5′ end identification of PF3D7_0617400 aslncRNA. (A) Schematic diagram of pT7SE-as0617400 and pT7-as0617400. The translation initial site of ATG of PF3D7_0617400 is marked as +1 and the stop codon is marked with +8375, the ∼2.5 kb var fragment (from +6412 to +3952) was amplified and inserted into pT7SE and pT7, respectively. The wavy line is the natural var aslncRNA. T7 pro, T7 promoter; T7 ter, T7 terminator. (B) 5′ end identification of PF3D7_0617400 aslncRNA. PCR products of 5′ end RACE obtained according to the standard manuals. M: the DNA marker λ-EcoT14 I digest (TAKARA), lane1: PCR products of 5′ RACE. (C) The PF3D7_0617400 aslncRNA transcriptional start sites. The translation start site of PF3D7_0617400 is marked as +1 on the schematic diagram. Capital letters are the coding sequence of the var exonI and the lower letters are intron sequence. The transcriptional start sites of the aslncRNAs are marked with underlined numbers and arrows. The underlined letters locations are displayed relative to translation start site, and the arrows indicate the aslncRNA transcription directions. (D) RT-qPCR quantification of the NLS-T7RNP transcription in C8/pT7- as0617400 (1) and C8/pT7SE-as0617400 (2). Relative transcripts numbers are normalized to serine-tRNA ligase gene (PF3D7_0717700). ∗P < 0.05 by paired two-tailed Student’s t-test. The RT-qPCR results are representative of three independent experiments with data indicating the mean +SD. (E) Western blot analysis of NLS-T7RNP expression in C8/pT7-as0617400 and C8/pT7SE-as0617400. The NLS-T7RNP is detected by anti-T7RNP antibody. Lane1: recombinant NLS-T7RNP expressed in E. coli BL21, Lane2: the wild type parasite 3D7 strain C8, Lane3: C8/pT7-as0617400, Lane4: C8/pT7SE-as0617400. (F) The subcellular localization of NLS-T7RNP in the transformant C8/pT7SE-as0617400 and C8/pT7-as0617400. NLS-T7RNP is marked with anti-Flag antibody (red) and is the nuclear marked with DAPI (blue). The scale bar is 25 μm.
The Detection of NLS-T7RNP Activity
To generate plasmid pUC15A, the replicon of pUC19 was replaced with the replicon p15A from pACYCDuet-1 (Novagen). Then the NLS-T7RNP gene was amplified from pCC4-NLS-T7RNP and inserted between HindIII and SphI sites of pUC15A, which named pUC15A-NLS-T7RNP. For the green fluorescent protein (GFP) expression, all of the E. coli containing plasmids were induced with 0.5 mM IPTG and cultured at 30°C for another 6 h after reaching OD600 of 0.8. Then GFP was detected by SDS-PAGE and Western blot. All the primers are listed in Supplementary Table S1.
Western Blot Analysis
The parasite samples were released from erythrocytes with 0.1% saponin, resuspended with 1% SDS/1 × PBS and sonicated for 5 min on high-level power by Bioruptor UCD-200 (Diagenode). After centrifuged at 1,3000 rpm for 2 min, the supernatant samples were separated by SDS-PAGE, and then were transferred to Immobilon-P transfer membranes (Millipore). Then each membrane of the samples were blocked with 5%BSA/1 × TBST (20 mM Tris-HCl, 0.9%NaCl, 0.1%Tween-20, 5%BSA, pH 7.5) and, respectively, incubated with anti-T7RNP (Novagen) and anti-Actin antibodies (Abmart). After being washed and incubated with anti-mouse IgG conjugated with HRP (Jackson ImmunoResearch Laboratories), the signal was developed with Immobilon Western Chemiluminescent HRP Substrate (Millipore) and imaged with Tanon 6200 System.
Immunofluorescence Assay
The immunofluorescence assay was performed to detect the subcellular localization of NLS-T7RNP in P. falciparum as described previously (Flueck et al., 2009). Parasites were harvested, resuspended with 1 × PBS, fixed by 4% paraformaldehyde (Electron Microscopy Sciences) at room temperature for 10 min and washed by 1 × PBS. Prepared samples were incubated with the primary antibody anti-Flag monoclonal antibody (1:1000, Abmart) and next with the secondary antibody anti-mouse IgG conjugated with DyLight 550 (1:2000, Thermo), and finally observed by Olympus IX73.
Localization of Induced and Dominant var mRNAs
The DNA templates of hybridization probes for RNA fluorescent in situ hybridization (RNA-FISH) were generated by PCR (Supplementary Table S1). The FISH probes of PF3D7_1240600 and PF3D7_0617400 mRNAs were, respectively, generated and labeled with biotin or fluorescein (Biotin-High Prime and Fluorescein-High Prime kits, Roche) according to each standard manual.
For detection of var gene mRNA localization, FISH was carried out as reported with some modifications (Epp et al., 2009). Fixed parasites for RNA-FISH were prepared as described above in the immunofluorescence assay and placed on polylysine-coated adhesion microscope slides (Citoglas). After permeabilization with 0.1% TritonX-100/1 × PBS for 7 min, washing with 1 × PBS and blocking with 1%BSA/1 × PBS for 30 min, FISH was performed in hybridization buffer [50% deionized formamide (Ambion), 10% dextran sulfate (MW > 500,000, Sigma-Aldrich), 2 × SSPE (Ambion), 250 μg/ml sheared salmon sperm DNA] with 70 ng/μl of each labeled probe at 37°C overnight. Slides were then washed with 50%formamide/2 × SSC and 2 × SSC (Ambion), blocked with 5%BSA/2 × SSC for 30 min, incubated with streptavidin-Alexa Fluor 594 (Thermo) for 30 min and washed with 2 × SSC. Finally, the results of RNA-FISH for PF3D7_1240600 and PF3D7_0617400 mRNAs were recorded by Olympus FV-1200.
Results
5′ End Identification of the var Gene aslncRNA
Published data demonstrated a positive correlation between active var gene and its aslncRNA (Jiang et al., 2013; Avraham et al., 2015), which was also repeated in this study. The dominant expressed var gene was identified as PF3D7_0617400 by RT-qPCR in P. falciparum NF54 strain A3 (Supplementary Figure S2A). The PF3D7_0617400 aslncRNA was also dominantly expressed among the selected var antisense transcripts in A3 strain (Supplementary Figure S2B), therefore, this strain was used for mapping 5′ end of the PF3D7_0617400 aslncRNA. To do so, total RNA of A3 strain was used, and a PCR fragment of ∼800 bp was obtained for sequencing (Figure 1B). Three transcriptional start sites of the PF3D7_0617400 aslncRNA were identified and mapped in the intron thymine-rich region, locating at 6776, 6745, and 6735 bp downstream of the translation start site (Figure 1C).
The var Gene Could Be Activated by Its aslncRNA
To identify the function of var aslncRNA, its template was cloned into the plasmids pT7SE and pT7. Because of the very high thymine content (73.9% in the first 176 bp) and short repeats at the 5′ end of PF3D7_0617400 aslncRNA (Figure 1C and Supplementary Figure S3), the first 364 bp of the PF3D7_0617400 aslncRNA template was unable to be obtained by PCR, so the artificial var aslncRNA was started from 6412 bp downstream of the PF3D7_0617400 start codon (Figure 1A).
The P. falciparum 3D7 strain C8 was, respectively, transfected with pT7-as0617400 or pT7SE-as0617400 and selected by BSD. As shown in the RT-qPCR results and western blot analysis, NLS-T7RNP was successfully transcribed and expressed in C8/pT7SE-as0617400 but not C8/pT7-as0617400 (Figures 1D,E). Furthermore, in the case of C8/pT7SE-as0617400, we expected to detect co-localization of the NLS-T7RNP with the nuclear DAPI staining in ring-stage and trophozoite-stage, which was also confirmed by immunofluorescence assay (Figure 1F). It indicated the proper and efficient transport of the protein into nucleus with the nuclear localization signal of Gal4p (Wittayacom et al., 2010), which helps NLS-T7RNP to function in the nucleus.
Next, total RNA of the tightly synchronized parasites C8/pT7SE-as0617400 and C8/pT7-as0617400 was harvested at 10–16 h post invasion and reverse transcribed into cDNA. The exogenous PF3D7_0617400 aslncRNA from episome in the transformants were detected by the RT-qPCR with specific primers p1/p2 (Supplementary Table S1). The gene PF3D7_1031000 (previously named pfs25), silenced in asexual blood stage, was chosen as a negative control (Dechering et al., 1999). Compared with PF3D7_1031000, the exogenous aslncRNA was transcribed successfully in C8/pT7SE-as0617400 but not in C8/pT7-as0617400 (Figures 2B,D), due to the lack of T7 RNA polymerase activity in C8/pT7-as0617400. Then the expression pattern of the var family was measured in both transformants. PF3D7_1240600, the dominant-expressed var in the wild type C8 strain (Supplementary Figure S2A), was still expressed in the two transformants (Figures 2A,C). Nevertheless, unlike the wild type and the negative control strain C8/pT7-as0617400, another var gene PF3D7_0617400 mRNA was found to be transcribed at a high level in C8/pT7SE-as0617400 (P < 0.05, Figure 2A). It seemed that the expression of the PF3D7_0617400 aslncRNA resulted in the activation of the corresponding silenced var gene.

Figure 2. The activation of silent var gene by its artificial aslncRNA. The expression of var family and artificial aslncRNA were detected in the transformant C8/pT7SE-as0617400 (A,B, respectively) and the negative control C8/pT7SE-as0617400 (C,D, respectively). PF3D7_0617400 was distinct expressed in C8/pT7SE-as0617400 but not C8/pT7-as0617400. ∗P < 0.05 by paired two-tailed Student’s t-test. (A–D) Relative transcripts numbers are normalized to serine-tRNA ligase gene (PF3D7_0717700). The RT-qPCR results are representative of three independent experiments with data indicating the mean+SD.
To confirm that the expression of PF3D7_0617400 was specifically activated by aslncRNA but not switched on in a long-term culture, pT7SE-as0617400 was transfected into G4 strain whose dominant var gene is PF3D7_0711700 (Supplementary Figure S2A). Similarly, PF3D7_0617400 was also detected to be active besides the dominant var PF3D7_0711700 (Supplementary Figure S4). These results indicated that the var gene aslncRNA could induce the corresponding var gene transcription.
Conserved TG Motif of var aslncRNA Is Not Essential for aslncRNA-Mediated var Activation
Previous studies revealed that the TG motifs were present in almost all the var gene introns and involved in var gene expression regulation (Avraham et al., 2012). The conserved TG motifs were also found in the intron region of PF3D7_0617400 aslncRNA (Supplementary Figure S3) but not in the exonI region of the artificial aslncRNA.
To identify whether the conserved TG motif was critical to aslncRNA-mediated var activation, the artificial PF3D7_0617400 aslncRNA template without the intron sequence was designed (Figure 3A) and expressed in the C8 strain. Then the transformant C8/pT7SE-as0617400-exonI was harvested at ring stage. As indicated in Figure 3B, the transcription of the artificial PF3D7_0617400 aslncRNA lacking TG motifs was detected in this transformant, and the aslncRNA also activated the corresponding var gene transcription at a high level compared with C8/pT7-as0617400 (P < 0.05, Figure 3C). The result showed that aslncRNA lacking the conserved motifs still retained the function of var activation. So that, the TG motif was not the essential element for var gene activation.
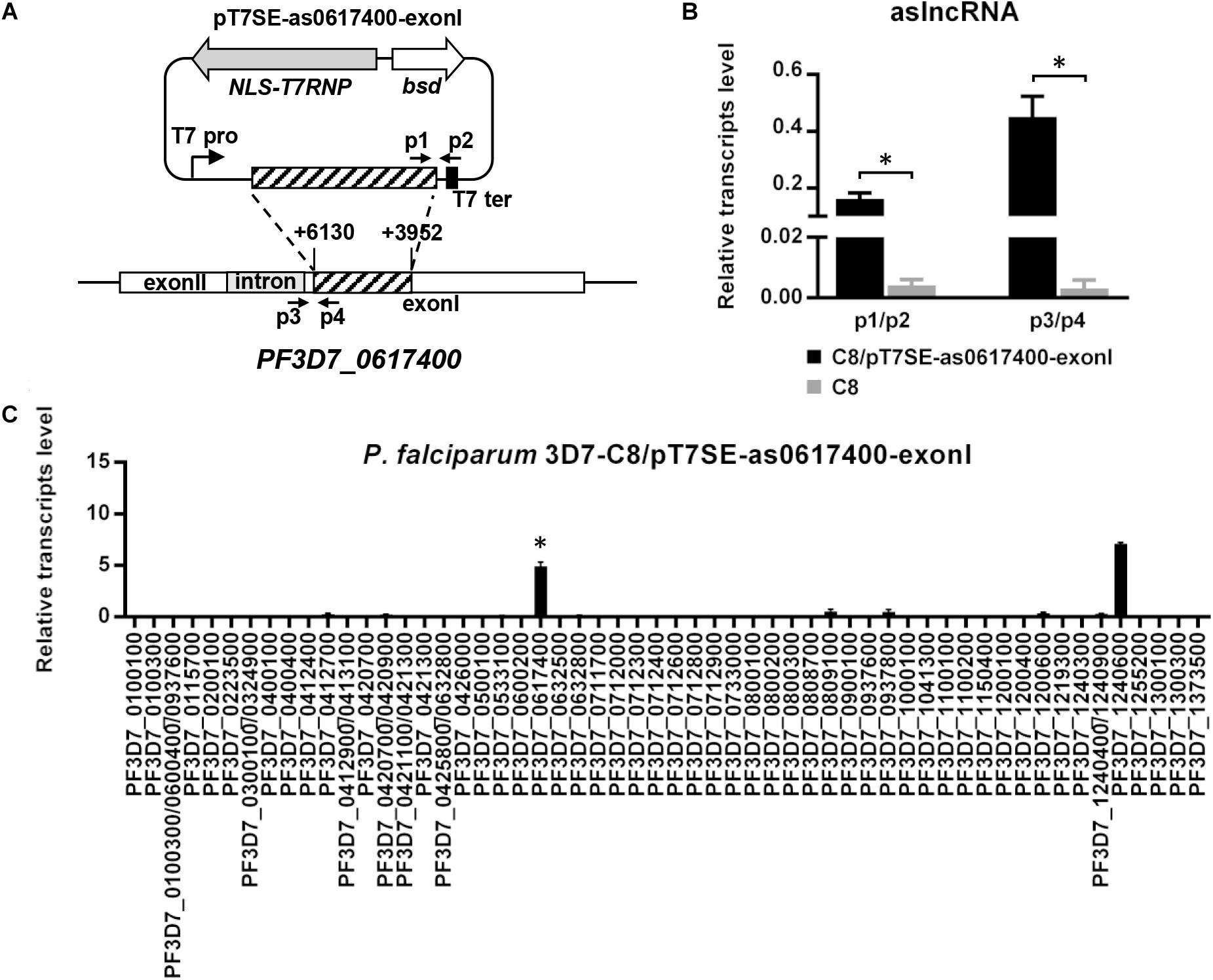
Figure 3. Silent var gene could be activated by its artificial aslncRNA lacking the intron sequence. (A) The schematic diagram of the episome for expressing the artificial PF3D7_0617400 aslncRNA lacking intron sequence. This plasmid was then used to transfected into C8 strain. Specific primer pairs p1/p2 and p3/p4 were used for the exogenous and endogenous artificial aslncRNA detection, respectively. T7 pro, T7 promoter; T7 ter, T7 terminator. (B) The expression of the exogenous and endogenous PF3D7_0617400 aslncRNAs in C8/pT7SE-as0617400-exonI (black) and C8 strain (gray). (C) The var gene expression pattern in C8/pT7SE-as0617400-exonI. In this transformant, the artificial PF3D7_0617400 aslncRNA was lacking intron region. PF3D7_0617400 was distinct expressed in C8/pT7SE-as0617400-exonI but not C8/pT7-as0617400. (B,C, respectively) Relative transcripts numbers are normalized to serine-tRNA ligase gene (PF3D7_0717700). The RT-qPCR results are representative of three independent experiments with data indicating the mean+SD. ∗P < 0.05 by paired two-tailed Student’s t-test.
The var aslncRNA Promoter in Intron Is Also Activated in the Induced var Gene
Some studies reported that the var gene and the corresponding aslncRNA were co-expressed (Jiang et al., 2013; Avraham et al., 2015). Thus, the exogenous aslncRNA activated the corresponding var gene, we decided to check if the aslncRNA promoter in the corresponding intron was also activated.
In order to test this, specific primer pairs for RT-PCR were prepared to distinguish the exogenous (p1/p2) and endogenous (p3/p4) transcripts in C8/pT7SE-as0617400-exonI (Figure 3A). The RT-qPCR data revealed the distinct transcription level of endogenous PF3D7_0617400 aslncRNA, whereas, no detectable expression of endogenous PF3D7_0617400 aslncRNA was found in the wild type C8 strain at the ring stage (Figure 3B). These results indicated that the PF3D7_0617400 intronic promoter was active to transcribe antisense transcripts when this var was induced by its corresponding artificial aslncRNA, and it further confirmed the co-activation of the promoters from the var gene and its intron. Regarding of the bi-directional promoter activities in the var gene intron, the sense lncRNA derived from the PF3D7_0617400 intron was also measured with specific RT-qPCR primers. However, a very low level of the sense lncRNA was detected in C8 strain at the same stage, suggesting the intronic promoter responsible for the sense lncRNA synthesis remained silent at the ring stage (Data not shown).
Dominant and Induced var Genes Are Transcribed in Single Parasite
The RT-qPCR data indicated that both of the dominant and induced var genes could be detected in the transformant population (Figures 2A, 3C, and Supplementary Figure S4A). We were then further checked whether the induced var mRNA was exclusively expressed or simultaneous expressed with the dominant var gene in single parasite.
Thus, RNA-FISH probes were prepared and applied to detect dominant and induced var mRNAs. In the wild type C8 strain, according to its var expression pattern, PF3D7_1240600 mRNA (red) was detected while the observation of PF3D7_0617400 mRNA (green) was failed (Figure 4A). In C8/pT7SE-as0617400, the PF3D7_0617400 mRNA (green) could be observed together with PF3D7_1240600 mRNA (red) in most parasite (102/116), and only 14 out of 116 parasites predominantly transcribed PF3D7_1240600 (Figures 4A,B). No parasite was found with a single green signal in C8/pT7SE-as0617400. This observation revealed the widespread co-expression of two var genes in one parasite. It suggested that the induced var gene by exogenous aslncRNA could get outside the MEE mechanism control and co-transcribed with previously dominant var gene in one cell.
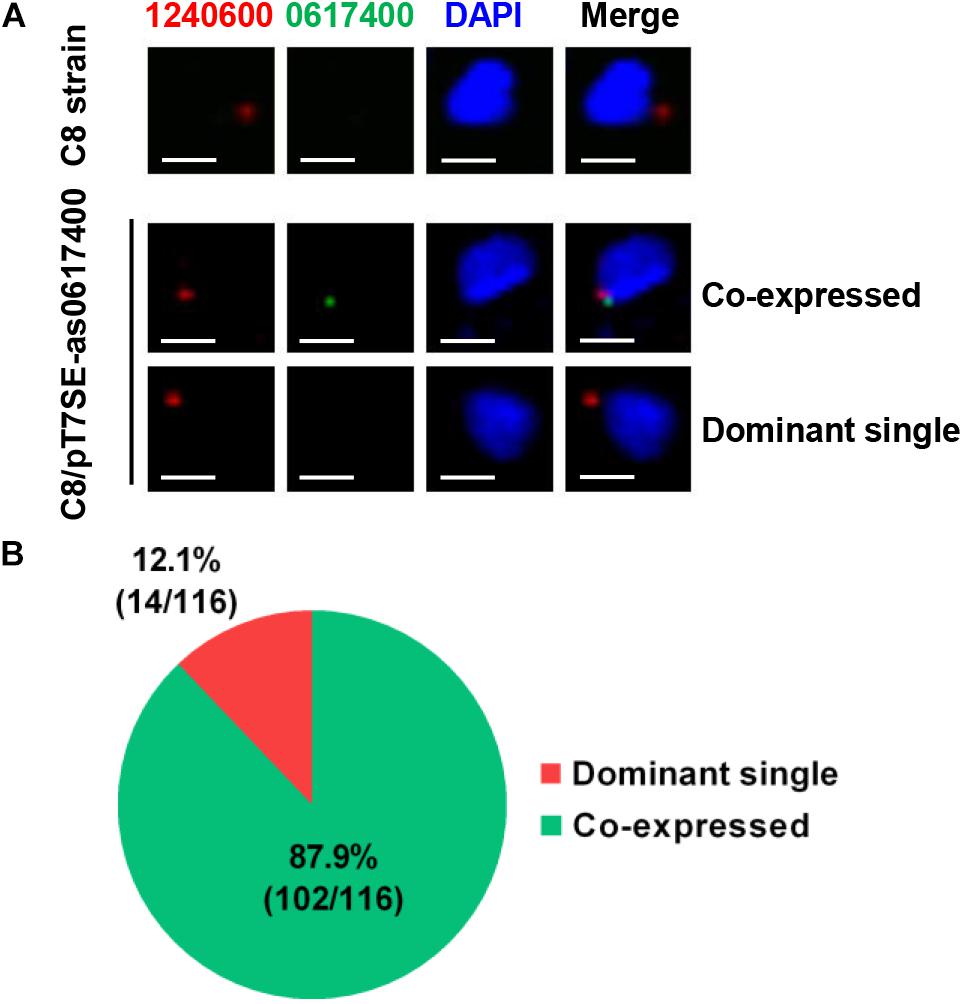
Figure 4. The subcellular location of var gene mRNAs by RNA-FISH. (A) The fluorescent microscopy images of the dominant and induced var mRNAs in the wild type C8 strain and the transformant C8/pT7SE- as0617400. The parasites were harvested at ring stage. The subcellular localization of the dominant PF3D7_1240600 (red) and induced var PF3D7_0617400 (green) were detected by RNA-FISH, the nucleus was stained with DAPI (blue). The scale bar is 2 μm. (B) In 116 parasites of C8/pT7SE-as0617400, PF3D7_0617400 mRNA (green) was observed together with PF3D7_1240600 mRNA (red) in 102 parasites, and only 14 parasites predominantly transcribed PF3D7_1240600.
In addition, it was noteworthy that the dominant PF3D7_1240600 expression level in C8/pT7SE-as0617400 was only half of that in the negative control (Figures 2A,C), which was probably caused by competitive transcription between the induced and dominant var genes in one parasite.
Discussion
It was described previously that the var aslncRNA co-expressed with its var gene (Jiang et al., 2013; Avraham et al., 2015). Nevertheless, it is disputable whether aslncRNA is involved in var transcription regulation. Therefore, the role of aslncRNA in var gene regulation is significant to be identified, which also will help to understand the var gene MEE mechanism. Remarkably, the var intron, as promoter of the var aslncRNA, was regarded to be involved in var expression regulation by potential cis–elements interactions with other factors (Calderwood et al., 2003; Voss et al., 2006; Dzikowski et al., 2007; Avraham et al., 2012). Therefore, it may disrupt functions or interactions of these factors by using genomic engineering strategy to modify the endogenous var gene intron. To avoid these interference, we investigated the var gene aslncRNA function via an episome introduced into P. falciparum. In such system, the genome is not modified and no additional intron sequence is introduced into the parasite, which should avoid disruptions or changes of some potential cis- or trans- elements interactions.
With T7 RNA polymerase system, the var aslncRNA is firstly identified as a functional activator to be responsible for the transcriptional regulation of corresponding var gene. In this study, the var gene PF3D7_0617400 is activated following the transcription of the additional aslncRNA from episome, showing that aslncRNA is a key activator on var gene transcription. Interestingly, PF3D7_0712600, which possess 73% identity with the exonI region of the PF3D7_0617400 aslncRNA template, failed to be active in all the transformants of C8/pT7SE-as0617400, C8/pT7SE-as0617400-exonI, and G4/pT7SE-as0617400 according to the RT-qPCR results (Figures 2A, 3C, and Supplementary Figure S4A). The data indicate that aslncRNA-mediated var activation is specific.
The result of RNA-FISH reveals that the previously dominant and newly induced var could be co-expressed in one parasite, and a competitive transcription between the dominant and induced var is also discovered in transformants (Figures 2A, 4). The induced var gene competes with the dominant var gene for some limited factors (e.g., transcriptional factors, chromatin remodeling factors and subnuclear compartment), which results in a transcription decrease of the latter. Although this status of co-expression potentially resembles to the transition state of the var gene switching, we could not manage to promote further development from co-existence to exclusive transcription of induced var by a long-term culture (Supplementary Figure S5). This implies other factors involved in the var MEE maintenance and development. Thus, we are more likely to regard the aslncRNA-mediated var activation not as the trigger but as a relative independent intermediary step of the var gene MEE regulation.
Generally, the long noncoding RNA (lncRNA) function in two ways: lncRNA–microRNA interaction or lncRNA–protein interaction. Mature microRNA is about 22-nucleotide RNA molecules in cytoplasm (Lund et al., 2004). If interacted with microRNA, lncRNA also should be exported into cytoplasm. The existing evidences demonstrated that P. falciparum var aslncRNA was located in cell nucleus (Epp et al., 2009). Therefore, we do not think that the var aslncRNA activates the corresponding var gene by interacting with microRNA. It is worth noting that the conserved TG motif, in all of the var intron region, has been demonstrated to be involved in var gene regulation. This element is merely found in the intron region of the var aslncRNA but not exonI region. In Avraham’ model, the TG motif (named “insulator-like PE” in their study, Avraham et al., 2015) bond with some silence-related factors and maintained the promoter–intron interaction, which made the var gene silence. The var aslncRNA could incorporate into chromatin in a sequence-specific manner and compete the silence-related factors with its TG motif, which resulted in the disruption of the promoter–intron interaction, then, the var promoter would be active. In our study, we also confirmed that var aslncRNA activatory function is sequence-dependent. However, we find that the var aslncRNA lacking the intron sequence still has capability of var activation (Figure 3C), which implies that the aslncRNA-mediated var activation is not by the way of binding with silence-related proteins with these conserved elements on the var aslnRNA.
Based on our data and Avraham’s study, we try to explain the mechanism underlying aslncRNA function: not only var but also non-var gene aslncRNA can specifically activate var gene promoter with its polymorphic sequence. As we know, Schmitz and collaborators found the DNA methyltransferase DNMT3b could be recruited by DNA-RNA triplex, and mediate CpG methylation to repress rRNA genes (Schmitz et al., 2010). Similarly, the lncRNA MEG3 and HOTAIR also can regulate genes by forming DNA-RNA triplex structure (Mondal et al., 2015; Kalwa et al., 2016). These studies revealed a novel regulation mechanism associated with lncRNA. According to these data and ours, we propose that there are potential binding domains in aslncRNA which could form the DNA-RNA triplex with the corresponding DNA. Then, the RNA-DNA triplex could further recruit factors to regulate the var transcription.
Author Contributions
LJ, QJ, and SL designed the experiments. QJ, LC, LZ, XC, and MS performed experiments. QJ, LC, LZ, XC, NG, XD, MS, and LJ analyzed data. QJ, LC, LJ, NG, XD, and LJ wrote the paper. All authors read, contributed feedback to, and approved the final manuscript.
Funding
This research was supported by the National Key R&D Program of China (2018YFA0507300), the National Science and Technology Major Project (2018ZX10101004003001), the National Natural Science Foundation of China (31571345, 31771455, and 81772218), the Chinese Academy of Sciences (GJHZ1703 and 153831WGZJTPYJY20170005), and by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health grant R01AI116466. All authors have reviewed and agreed upon the content of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Xiangfei Song for kindly providing the plasmid pET28a-GFP and we also gratefully thank Zhiyun Gao for her critical comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03117/full#supplementary-material
References
Avraham, I., Pozner, G., Eshar, S., Fastman, Y., Kolevzon, N., Yavin, E., et al. (2015). Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 112, E982–E991. doi: 10.1073/pnas.1420855112
Avraham, I., Schreier, J., and Dzikowski, R. (2012). Insulator-like pairing elements regulate silencing and mutually exclusive expression in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 109, E3678–E3686. doi: 10.1073/pnas.1214572109
Baum, J., Papenfuss, A. T., Mair, G. R., Janse, C. J., Vlachou, D., Waters, A. P., et al. (2009). Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 37, 3788–3798. doi: 10.1093/nar/gkp239
Bryant, J. M., Regnault, C., Scheidig-Benatar, C., Baumgarten, S., Guizetti, J., and Scherf, A. (2017). CRISPR/Cas9 genome editing reveals that the intron is not essential for var2csa gene activation or silencing in Plasmodium falciparum. mBio 8:e729-717. doi: 10.1128/mbio.00729-17
Calderwood, M. S., Gannoun-Zaki, L., Wellems, T. E., and Deitsch, K. W. (2003). Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 278, 34125–34132. doi: 10.1074/jbc.m213065200
Cranmer, S. L., Magowan, C., Liang, J., Coppel, R. L., and Cooke, B. M. (1997). An alternative to serum for cultivation of Plasmodium falciparum in vitro. Trans. R. Soc. Trop. Med. Hyg. 91, 363–365. doi: 10.1016/S0035-9203(97)90110-3
Dechering, K. J., Kaan, A. M., Mbacham, W., Wirth, D. F., Eling, W., Konings, R. N., et al. (1999). Isolation and functional characterization of two distinct sexual-stage-specific promoters of the human malaria parasite Plasmodium falciparum. Mol. Cell Biol. 19, 967–978. doi: 10.1128/mcb.19.2.967
Deitsch, K. W., Calderwood, M. S., and Wellems, T. E. (2001). Malaria: cooperative silencing elements in var genes. Nature 412, 875–876. doi: 10.1038/35091146
Deitsch, K. W., Lukehart, S. A., and Stringer, J. R. (2009). Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat. Rev. Microbiol. 7, 493–503. doi: 10.1038/nrmicro2145
Duraisingh, M. T., Voss, T. S., Marty, A. J., Duffy, M. F., Good, R. T., Thompson, J. K., et al. (2005). Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121, 13–24. doi: 10.1016/j.cell.2005.01.036
Dzikowski, R., Li, F., Amulic, B., Eisberg, A., Frank, M., Patel, S., et al. (2007). Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 8, 959–965. doi: 10.1038/sj.embor.7401063
Dzikowski, R., Templeton, T. J., and Deitsch, K. (2006). Variant antigen gene expression in malaria. Cell. Microbiol. 8, 1371–1381. doi: 10.1111/j.1462-5822.2006.00760.x
Epp, C., Li, F., Howitt, C. A., Chookajorn, T., and Deitsch, K. W. (2009). Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15, 116–127. doi: 10.1261/rna.1080109
Flueck, C., Bartfai, R., Volz, J., Niederwieser, I., Salcedo-Amaya, A. M., Alako, B. T., et al. (2009). Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 5:e1000569. doi: 10.1371/journal.ppat.1000569
Fraschka, S. A.-K., Henderson, R. W. M., and Bártfai, R. (2016). H3.3 demarcates GC-rich coding and subtelomeric regions and serves as potential memory mark for virulence gene expression in Plasmodium falciparum. Sci. Rep. 6:31965. doi: 10.1038/srep31965
Jiang, L., Mu, J., Zhang, Q., Ni, T., Srinivasan, P., Rayavara, K., et al. (2013). PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499, 223–227. doi: 10.1038/nature12361
Kalwa, M., Hänzelmann, S., Otto, S., Kuo, C. C., Franzen, J., Joussen, S., et al. (2016). The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 44, 10631–10643. doi: 10.1093/nar/gkw802
Leech, J. H., Barnwell, J. W., Miller, L. H., and Howard, R. J. (1984). Identification of a strain-specific malarial antigen exposed on the surface of Plasmodium falciparum-infected erythrocytes. J. Exp. Med. 159, 1567–1575. doi: 10.1084/jem.159.6.1567
Lopez-Rubio, J. J., Gontijo, A. M., Nunes, M. C., Issar, N., Hernandez Rivas, R., and Scherf, A. (2007). 5 flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 66, 1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x
Lund, E., Güttinger, S., Calado, A., Dahlberg, J. E., and Kutay, U. (2004). Nuclear export of microRNA precursors. Science 303, 95–98. doi: 10.1126/science.1090599
Maier, A. G., Braks, J. A., Waters, A. P., and Cowman, A. F. (2006). Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Mol. Biochem. Parasitol. 150, 118–121. doi: 10.1016/j.molbiopara.2006.06.014
Mondal, T., Subhash, S., Vaid, R., Enroth, S., Uday, S., Reinius, B., et al. (2015). MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat. Commun. 6:7743. doi: 10.1038/ncomms8743
Pays, E., Vanhamme, L., and Perez-Morga, D. (2004). Antigenic variation in Trypanosoma brucei: facts, challenges and mysteries. Curr. Opin. Microbiol. 7, 369–374. doi: 10.1016/j.mib.2004.05.001
Prucca, C. G., Slavin, I., Quiroga, R., Elías, E. V., Rivero, F. D., Saura, A., et al. (2008). Antigenic variation in Giardia lamblia is regulated by RNA interference. Nature 456, 750–754. doi: 10.1038/nature07585
Ralph, S. A., Bischoff, E., Mattei, D., Sismeiro, O., Dillies, M. A., Guigon, G., et al. (2005a). Transcriptome analysis of antigenic variation in Plasmodium falciparum-var silencing is not dependent on antisense RNA. Genome Biol. 6:R93.
Ralph, S. A., Scheidig-Benatar, C., and Scherf, A. (2005b). Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. Sci. U.S.A. 102, 5414–5419. doi: 10.1073/pnas.0408883102
Rowe, A., Obeiro, J., Newbold, C. I., and Marsh, K. (1995). Plasmodium falciparum rosetting is associated with malaria severity in Kenya. Infect. Immun. 63, 2323–2326.
Rug, M., and Maier, A. G. (2012). Transfection of Plasmodium falciparum in Malaria. Methods Mol. Biol. 923, 75–98. doi: 10.1007/978-1-62703-026-7_6
Salanti, A., Staalsoe, T., Lavstsen, T., Jensen, A. T., Sowa, M. P., Arnot, D. E., et al. (2003). Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 49, 179–191.
Scherf, A., Hernandez-Rivas, R., Buffet, P., Bottius, E., Benatar, C., Pouvelle, B., et al. (1998). Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 17, 5418–5426. doi: 10.1093/emboj/17.18.5418
Schmitz, K.-M., Mayer, C., Postepska, A., and Grummt, I. (2010). Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269. doi: 10.1101/gad.590910
Volz, J. C., Bártfai, R., Petter, M., Langer, C., Josling, G. A., Tsuboi, T., et al. (2012). PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe 11, 7–18. doi: 10.1016/j.chom.2011.11.011
Voss, T. S., Healer, J., Marty, A. J., Duffy, M. F., Thompson, J. K., Beeson, J. G., et al. (2006). A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 439, 1004–1008. doi: 10.1038/nature04407
Voss, T. S., Kaestli, M., Vogel, D., Bopp, S., and Beck, H.-P. (2003). Identification of nuclear proteins that interact differentially with Plasmodium falciparum var gene promoters. Mol. Microbiol. 48, 1593–1607. doi: 10.1046/j.1365-2958.2003.03528.x
Keywords: malaria, Plasmodium falciparum, var gene, long non-coding RNA, T7 RNA polymerase
Citation: Jing Q, Cao L, Zhang L, Cheng X, Gilbert N, Dai X, Sun M, Liang S and Jiang L (2018) Plasmodium falciparum var Gene Is Activated by Its Antisense Long Noncoding RNA. Front. Microbiol. 9:3117. doi: 10.3389/fmicb.2018.03117
Received: 16 April 2018; Accepted: 03 December 2018;
Published: 18 December 2018.
Edited by:
Wei Hu, Fudan University, ChinaReviewed by:
Abhisheka Bansal, Jawaharlal Nehru University, IndiaBang Shen, Huazhong Agricultural University, China
Xing He, Second Military Medical University, China
Copyright © 2018 Jing, Cao, Zhang, Cheng, Gilbert, Dai, Sun, Liang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingqing Jing, qqjing@ips.ac.cn Shaohui Liang, lsh@wmu.edu.cn Lubin Jiang, lbjiang@ips.ac.cn
†These authors have contributed equally to this work
 Qingqing Jing1,2*†
Qingqing Jing1,2*† Liangliang Zhang
Liangliang Zhang Nicolas Gilbert
Nicolas Gilbert Shaohui Liang
Shaohui Liang Lubin Jiang
Lubin Jiang