TRIM71 Deficiency Causes Germ Cell Loss During Mouse Embryogenesis and Is Associated With Human Male Infertility
- 1Life and Medical Sciences Institute, University of Bonn, Bonn, Germany
- 2Institute of Reproductive Genetics, University of Münster, Münster, Germany
- 3Institute of Medical Informatics, University of Münster, Münster, Germany
- 4Institute of Pathology, University Hospital Bonn, Bonn, Germany
- 5Institute for Veterinary Anatomy, Histology and Embryology, Justus Liebig University Gießen, Gießen, Germany
- 6Hessian Centre of Reproductive Medicine (HZRM), Justus Liebig University Gießen, Gießen, Germany
- 7Department of Human Genetics, Radboud University Medical Center, Nijmegen, Netherlands
- 8Centre of Reproductive Medicine and Andrology, Institute of Reproductive and Regenerative Biology, University Hospital Münster, Münster, Germany
- 9Centre of Reproductive Medicine and Andrology, Department of Clinical and Surgical Andrology, University Hospital Münster, Münster, Germany
- 10Institute of Experimental Oncology, University Hospital Bonn, Bonn, Germany
Mutations affecting the germline can result in infertility or the generation of germ cell tumors (GCT), highlighting the need to identify and characterize the genes controlling germ cell development. The RNA-binding protein and E3 ubiquitin ligase TRIM71 is essential for embryogenesis, and its expression has been reported in GCT and adult mouse testes. To investigate the role of TRIM71 in mammalian germ cell embryonic development, we generated a germline-specific conditional Trim71 knockout mouse (cKO) using the early primordial germ cell (PGC) marker Nanos3 as a Cre-recombinase driver. cKO mice are infertile, with male mice displaying a Sertoli cell-only (SCO) phenotype which in humans is defined as a specific subtype of non-obstructive azoospermia characterized by the absence of germ cells in the seminiferous tubules. Infertility in male Trim71 cKO mice originates during embryogenesis, as the SCO phenotype was already apparent in neonatal mice. The in vitro differentiation of mouse embryonic stem cells (ESCs) into PGC-like cells (PGCLCs) revealed reduced numbers of PGCLCs in Trim71-deficient cells. Furthermore, TCam-2 cells, a human GCT-derived seminoma cell line which was used as an in vitro model for PGCs, showed proliferation defects upon TRIM71 knockdown. Additionally, in vitro growth competition assays, as well as proliferation assays with wild type and CRISPR/Cas9-generated TRIM71 mutant NCCIT cells showed that TRIM71 also promotes proliferation in this malignant GCT-derived non-seminoma cell line. Importantly, the PGC-specific markers BLIMP1 and NANOS3 were consistently downregulated in Trim71 KO PGCLCs, TRIM71 knockdown TCam-2 cells and TRIM71 mutant NCCIT cells. These data collectively support a role for TRIM71 in PGC development. Last, via exome sequencing analysis, we identified several TRIM71 variants in a cohort of infertile men, including a loss-of-function variant in a patient with an SCO phenotype. Altogether, our work reveals for the first time an association of TRIM71 deficiency with human male infertility, and uncovers further developmental roles for TRIM71 in the germline during mouse embryogenesis.
Introduction
Despite the recent insights into the regulation of mammalian germ cell development and the advances in assisted reproductive technologies, infertility remains a common problem in modern society affecting ∼15% of couples in industrialized countries (Agarwal et al., 2015). Genetic defects during germ cell development can drive infertility and increase susceptibility to germ cell tumors (GCT) (Raman et al., 2005; Looijenga et al., 2010; Zorrilla and Yatsenko, 2013).
Primordial germ cells (PGCs) are the first established germ cell population during embryonic development. In mice, PGC specification is initiated in the post-implantation epiblast at embryonic day (E) 6.25 in response to BMP4 signaling from the extra-embryonic ectoderm (Lawson et al., 1999). BMP4 activates the expression of Prdm14 (Yamaji et al., 2008) and Prdm1/Blimp1 (Ohinata et al., 2005). BLIMP1 then activates the expression of Tfap2c (Kurimoto et al., 2008), and together BLIMP1, TFAP2C and PRDM14 establish the transcriptional program required for PGC specification (Wang and Cao, 2016) which is completed at E7.5. At this time point, about 40 cells in the proximal epiblast express early PGC-specific markers such as Nanos3 (Ginsburg et al., 1990; Tsuda et al., 2003). PGCs then migrate to the genital ridges (developing gonads) while slowly proliferating (Tam and Snow, 1981; Anderson et al., 2000). At E10.5, around 1000 PGCs colonize the genital ridges, upregulate late PGC markers such as Ddx4/Vasa (Fujiwara et al., 1994; Tanaka et al., 2000) and Gcna1 (Enders and May, 1994), and undergo significant proliferation before initiating sex determination (Bowles and Koopman, 2010; Ewen and Koopman, 2010). Importantly, PGCs that fail to reach the genital ridges or to further differentiate into gametes are the origin of GCT, which often affect children and young adults (Raman et al., 2005; Looijenga et al., 2010; Zorrilla and Yatsenko, 2013; Pierce et al., 2018). Therefore, unraveling the processes controlling germline development will not only contribute to our understanding of infertility, but may also offer unique opportunities for the treatment of GCT.
Tripartite Motif Containing 71 (TRIM71) belongs to the TRIM-NHL protein family. TRIM71’s versatile domain structural organization (Ecsedi and Grosshans, 2013) enables its role as an E3 ubiquitin ligase (Chen et al., 2012; Nguyen et al., 2017; Ren et al., 2018; Hu et al., 2019) and an mRNA-binding protein (Chang et al., 2012; Loedige et al., 2013; Worringer et al., 2014; Mitschka et al., 2015; Torres-Fernández et al., 2019). TRIM71 function is essential for embryogenesis and its expression is mostly restricted to undifferentiated cells during early proliferative developmental stages, being downregulated in the course of differentiation (Schulman et al., 2005, 2008; Rybak et al., 2009; Cuevas et al., 2015). However, a postnatal Trim71 expression has also been observed in adult mouse testes (Rybak et al., 2009; Du et al., 2020) as well as in several GCT-derived cell lines (Rybak et al., 2009; Chang et al., 2012; Torres-Fernández et al., 2019), and a recent study has reported a postnatal function for TRIM71 in adult mouse spermatogenesis (Du et al., 2020). Furthermore, RNA sequencing of Trim71-deficient mouse embryonic stem cells (ESCs) revealed a decreased expression of genes associated with reproductive processes (Mitschka et al., 2015), suggesting an early developmental role for TRIM71 in the germline.
To elucidate the role of TRIM71 in mammalian embryonic germ cell development, we generated a mouse model with an early germline-specific depletion of Trim71 driven by the Nanos3 promoter (Nanos3-Cre). Additionally, we employed an in vitro approach for the differentiation of wild type and Trim71-deficient murine ESCs into PGC-like cells (PGCLCs) in order to study the role of TRIM71 during PGC specification. Furthermore, we used the human GCT-derived seminoma cell line TCam-2 as a surrogate in vitro model of PGCs and evaluated cell proliferation upon TRIM71 knockdown. Additionally, we used the human GCT-derived non-seminoma cell line NCCIT to generate TRIM71 mutations via CRISPR/Cas9 in order to investigate the role of TRIM71 in the proliferation of pluripotent embryonal carcinoma cells. Last, we used exome sequencing data from infertile men and developed a novel software tool named Haystack to search for novel genetic causes of infertility. Our work here identifies TRIM71 as a novel gene associated with human male infertility and uncovers a novel role for TRIM71 in the embryonic development of the germline.
Results
Trim71 Is Expressed in Spermatogonial Stem Cells and Is Essential for Mouse Fertility
Mice carrying a Trim71 homozygous deletion (KO, Trim71–/–) die during embryonic development (Supplementary Figure 1A; Schulman et al., 2008; Cuevas et al., 2015). In contrast, Trim71 heterozygous mice (HET, Trim71fl/–) are viable and fertile, although significantly smaller in length and weight than wild type (WT, Trim71fl/fl) littermates (Supplementary Figures 1B,C). We detected Trim71 expression in the testes – but not in other organs such as heart or kidney – of wild type and heterozygous adult mice, with Trim71 mRNA levels decreased in the testes of heterozygous mice (Figure 1A). We then measured the weight of several organs relative to the total body weight, and observed that testes – but neither heart nor kidney – were significantly smaller in heterozygous males compared to wild type males (Figure 1B). Accordingly, sperm counts were significantly reduced in adult heterozygous males (Figure 1C). These data suggested that Trim71 expression levels are important for male gonad development and revealed a haploinsufficiency of Trim71 in mice.
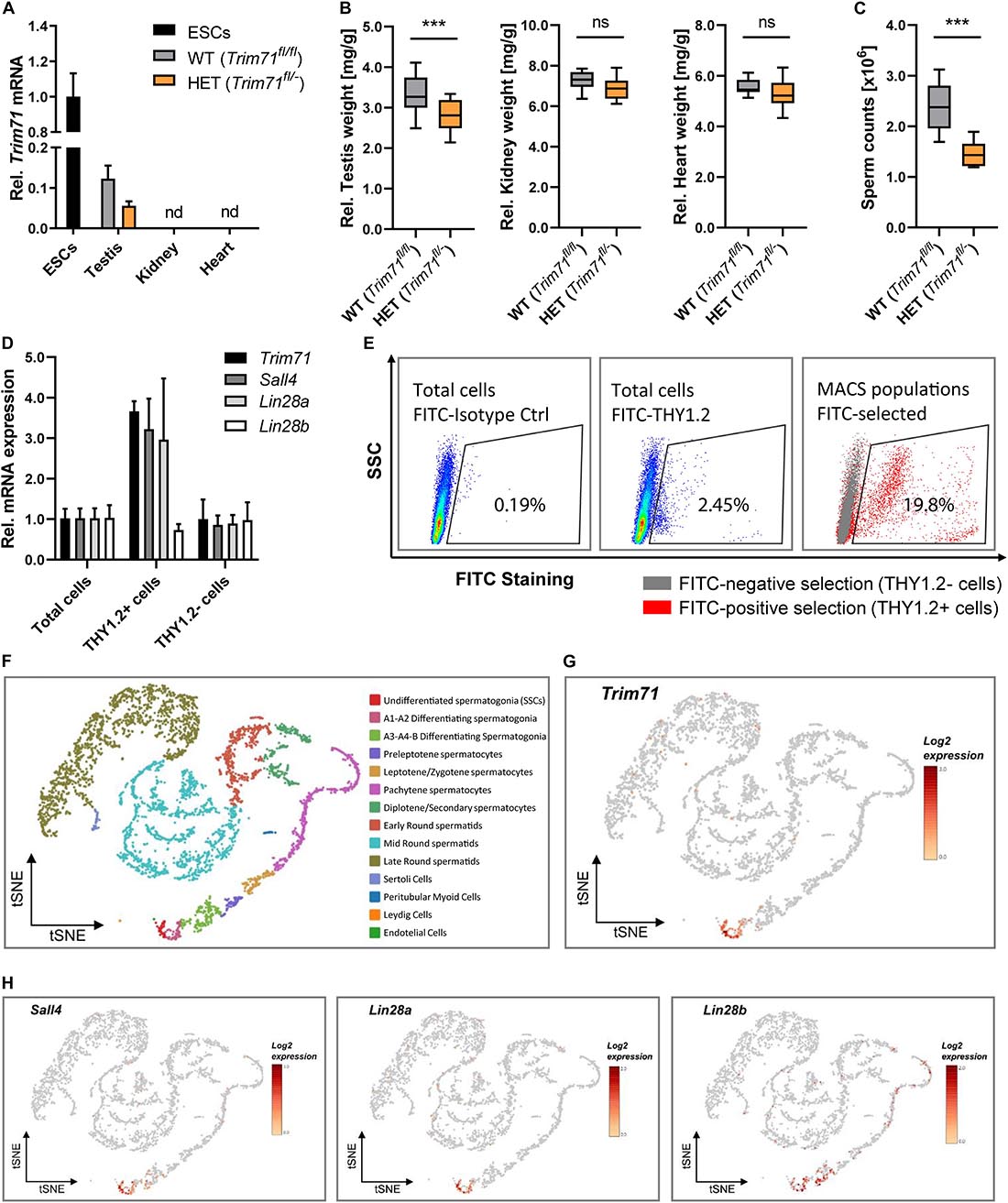
Figure 1. Trim71 is expressed in spermatogonial stem cells (SSCs) of adult fertile mice. (A) qRT-PCR of Trim71 relative to Hprt housekeeping gene in testis, kidney and heart of wild type (WT, Trim71fl/fl) and Trim71 heterozygous (HET, Trim71fl/−) adult male mice (10–14 weeks old), normalized to Trim71 relative levels in wild type murine ESCs (nd, not detected). Error bars represent SD (n = 3). (B) Organ weight [mg] relative to total body weight [g] of testis, kidney and heart of wild type (WT, Trim71fl/fl) and Trim71 heterozygous (HET, Trim71fl/−) male adult mice (10–14 weeks old). Graphs represent Tukey plots (n = 14–16). ***P-value < 0.005, ns, non-significant (unpaired Student’s t-test). (C) Epididymal sperm counts in wild type (WT, Trim71fl/fl) and Trim71 heterozygous (HET, Trim71fl/−) male adult mice (10–14 weeks old). ***P-value < 0.005 (unpaired Student’s t-test). (D) qRT-PCR of Trim71, the SSC markers Sall4 and Lin28a, and the differentiating spermatogonia marker Lin28b, relative to Hprt housekeeping gene in testes cell suspension before (total cells) and after THY1.2 MACS (THY1.2+/− cells). Error bars represent SD (n = 3). (E) Representative flow cytometry scatter plots of testes cell suspension before (total cells) and after THY1.2 MACS (THY1.2+/− cells). (F) tSNE plot (t-distributed stochastic neighbor embedding) of single-cell transcriptome data (scRNA-seq) from mouse testes as published by Hermann et al. (2018). Each dot represents a single cell and is colored according to its cluster identity as indicated on the figure key. (G) Expression pattern of Trim71 and (H) Sall4, Lin28a and Lin28b projected on the tSNE plot of the mouse scRNA-seq dataset. Red indicates high expression and gray indicates low or no expression, as shown by the figure key. See also Supplementary Figures 1, 2.
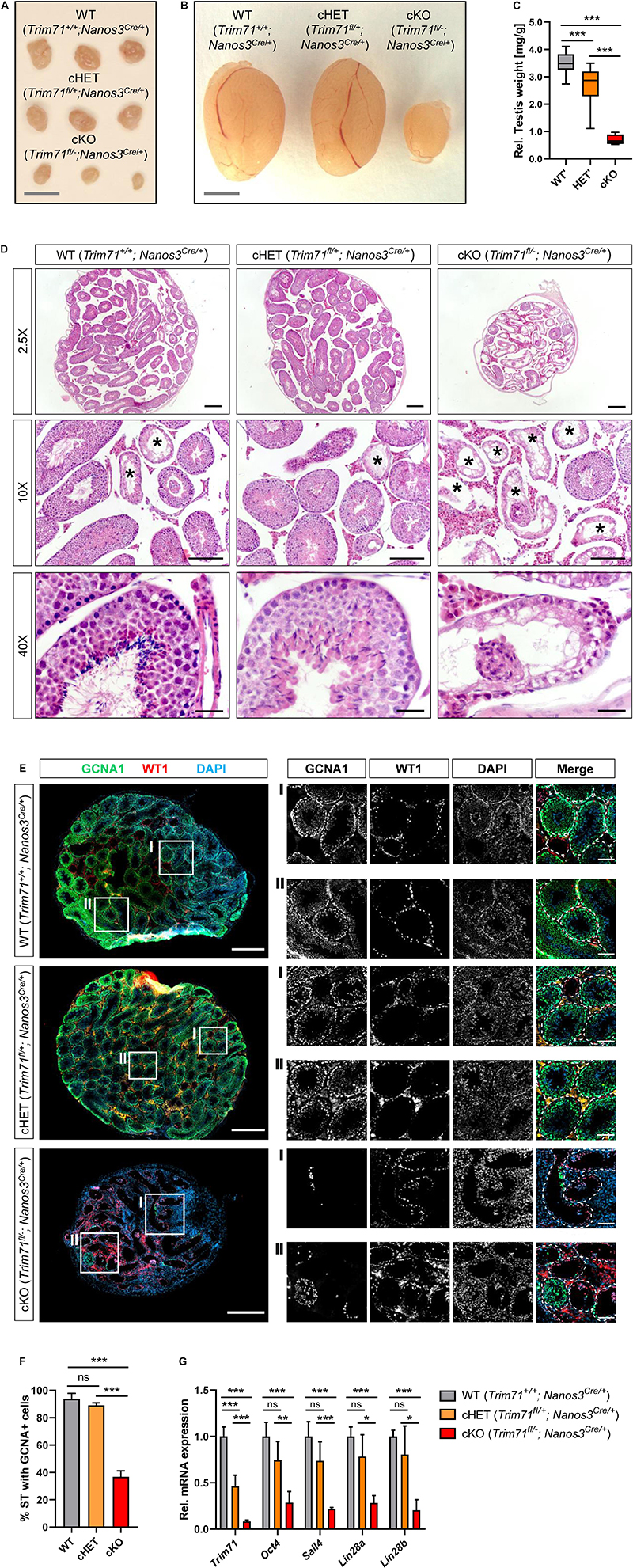
Figure 2. Germline-specific Trim71 cKO male mice present a Sertoli cell-only (SCO) phenotype. (A) Representative images of ovaries and (B) testes of adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) mice. Scale bars represent 2 mm. (C) Testis weight [mg] relative to total body weight [g] of wild type (WT’), Trim71 heterozygous (HET’) and germline-specific Trim71 knockout (cKO) mice, summarized from Supplementary Figure 2C, joining several Nanos3 genotypes per Trim71 genotype. Error bars represent SD (n = 8–29). ***P-value < 0.005 (one-way ANOVA, Tukey’s test). (D) Representative H&E stainings on paraffin testes cross-sections from adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) mice. Seminiferous tubules with a defective morphology are marked with an asterisk (*) in 10x magnification images. Scale bars represent 200, 100, and 20 μm in 2.5×, 10×, and 40× magnifications, respectively. (E) Representative immunofluorescence stainings on testes cryosections from adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) mice. Images show co-staining with GCNA1 (germ cells), WT1 (Sertoli cells) and DAPI (nuclei). For each genotype two regions – indicated as I and II – are depicted in higher magnification. Scale bars represent 500 μm for complete testes cross-sections and 100 μm for the magnification images. (F) Quantification of seminiferous tubules (ST) containing GCNA+ cells per testis cross-section from E, depicted as percentages. Error bars represent SD (n = 3). ***P-value < 0.005, ns, non-significant (unpaired Student’s t-test). (G) qRT-PCR of Trim71, the murine SSC markers Sall4, Lin28a and Oct4, and the differentiating spermatogonia marker Lin28b, relative to Hprt housekeeping gene in whole testis RNA of wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) male adult mice. Error bars represent SD (n = 3−6). ***P-value < 0.005; ***P-value < 0.01; *P-value < 0.05; ns, non-significant (unpaired Student’s t-test). See also Supplementary Figures 3, 4.
Since TRIM71 is usually expressed in undifferentiated cells, we hypothesized that its expression in mice testes is present in undifferentiated spermatogonia, also known as spermatogonial stem cells (SSCs). To validate this hypothesis, we used wild type testes cell suspensions to enrich SSCs (THY1.2 +) via magnetic-activated cell sorting (MACS) (Liao et al., 2016). We found an increase of Trim71 mRNA as well as the mRNAs of the known murine SSC-specific markers Sall4 (Gassei and Orwig, 2013) and Lin28a (Zheng et al., 2009) – but not the differentiating spermatogonia marker Lin28b (Gaytan et al., 2013) – in the THY1.2 + enriched cell population (Figures 1D,E). This result indicated that Trim71 expression in adult mouse testes is enriched in SSCs. Indeed, single-cell RNA sequencing analysis of mouse and human adult testicular tissue confirmed a restricted expression of Trim71 in SSCs (Figures 1F–H and Supplementary Figure 2; Hermann et al., 2018).
To study the function of TRIM71 in male gonad development, a germline-specific Trim71 conditional knockout mouse (cKO) was generated with the Cre recombinase expressed under the promoter of the early PGC marker Nanos3 (Suzuki et al., 2008; Supplementary Figure 3A). For a first functional evaluation, adult cKO mice (Trim71fl/–; Nanos3Cre/+) were crossed with wild type (Trim71+/+) mice. Trim71 deficiency in the germline results in infertility in both sexes, as neither cKO males nor females were able to produce offspring after numerous mating attempts over the course of several months (Supplementary Figure 3B).
Germline-Specific Trim71 cKO Male Mice Display a Sertoli Cell-Only (SCO)-Like Phenotype
Macroscopic analysis of cKO male and female reproductive organs revealed a dramatic reduction in testis and ovary size, respectively (Figures 2A–C and Supplementary Figure 3C). Hematoxylin and eosin (H&E) staining of adult male testis cross-sections showed that most seminiferous tubules in cKO testes were lacking signs of spermatogenesis (Figure 2D). Furthermore, immunofluorescence staining of germ cells (GCNA1 Koshimizu et al., 1995; Tanaka et al., 1998) and Sertoli cells (WT1 Mundlos et al., 1993) showed a dramatic reduction of GCNA + cells in cKO mice (Figures 2E,F). Accordingly, the expression of murine SSC-specific markers Oct4, Sall4 and Lin28a, as well as the differentiating spermatogonia marker Lin28b, was dramatically reduced in whole testis RNA of cKO mice (Figure 2G), representing the deficit of germ cells in these mice.
This phenotype is reminiscent of a human condition known as Sertoli cell-only (SCO) phenotype, which is a specific type of non-obstructive azoospermia characterized by a total or substantial absence of germ cells in the seminiferous tubules, and whose causes remain mostly unknown (Tüttelmann et al., 2011, 2018; Koc et al., 2019). Our results here suggest that Trim71 cKO male mice display an SCO-like phenotype with most seminiferous tubules lacking germ cells.
Infertility in Germline-Specific Trim71 cKO Male Mice Has an Embryonic Origin
To investigate the origin of infertility in Trim71 cKO mice, we first analyzed available datasets for the expression of Trim71 in murine male and female gonads. While Trim71 expression in males is present in both fetal and adult testes, Trim71 expression in females is detected in fetal ovaries but not in adult ovaries (Supplementary Figure 4A; Han et al., 2018). These results were confirmed by western blot comparing TRIM71 expression in adult testes and ovaries of wild type and Trim71 cKO mice (Supplementary Figure 4B). Such an expression pattern may indicate that TRIM71 fulfills embryonic and postnatal functions in the male germline, but only embryonic functions in the female germline. Because a postnatal function of TRIM71 in adult mouse spermatogenesis has already been described (Du et al., 2020), we decided to focus on the possible TRIM71 function during embryonic germline development. As Nanos3 expression in mice embryos is detected as early as E7.5 during PGC specification (Tsuda et al., 2003), the infertility observed in both male and female Trim71 cKO mice may reflect a function of TRIM71 in an early stage of germ cell development. Thus, we next analyzed available datasets for Trim71 expression in murine fetal gonads. The expression of Trim71 followed a similar pattern for male and female gonads during different mouse developmental stages (Supplementary Figure 4C; Soh et al., 2015; Zhao et al., 2018) which correlated with Trim71 expression in isolated male and female germ cells at the same stages (Supplementary Figure 4D; Seisenberger et al., 2012). This expression pattern supports a possible role for TRIM71 in germ cell development. In order to determine whether the deficit of germ cells observed in adult Trim71 cKO male mice derives from defects during embryonic development, we conducted H&E staining and GCNA1-WT1 immunostaining on testis cross-sections from neonatal (P0.5) male mice (Figures 3A,B). Indeed, the SCO-like phenotype previously observed in adult male cKO mice was already apparent in neonatal cKO mice as shown by a reduction in gonocyte-containing seminiferous tubules (Figures 3B,C). These results confirm that Trim71 expression during embryonic development is required for the generation and/or maintenance of the male germline before birth.
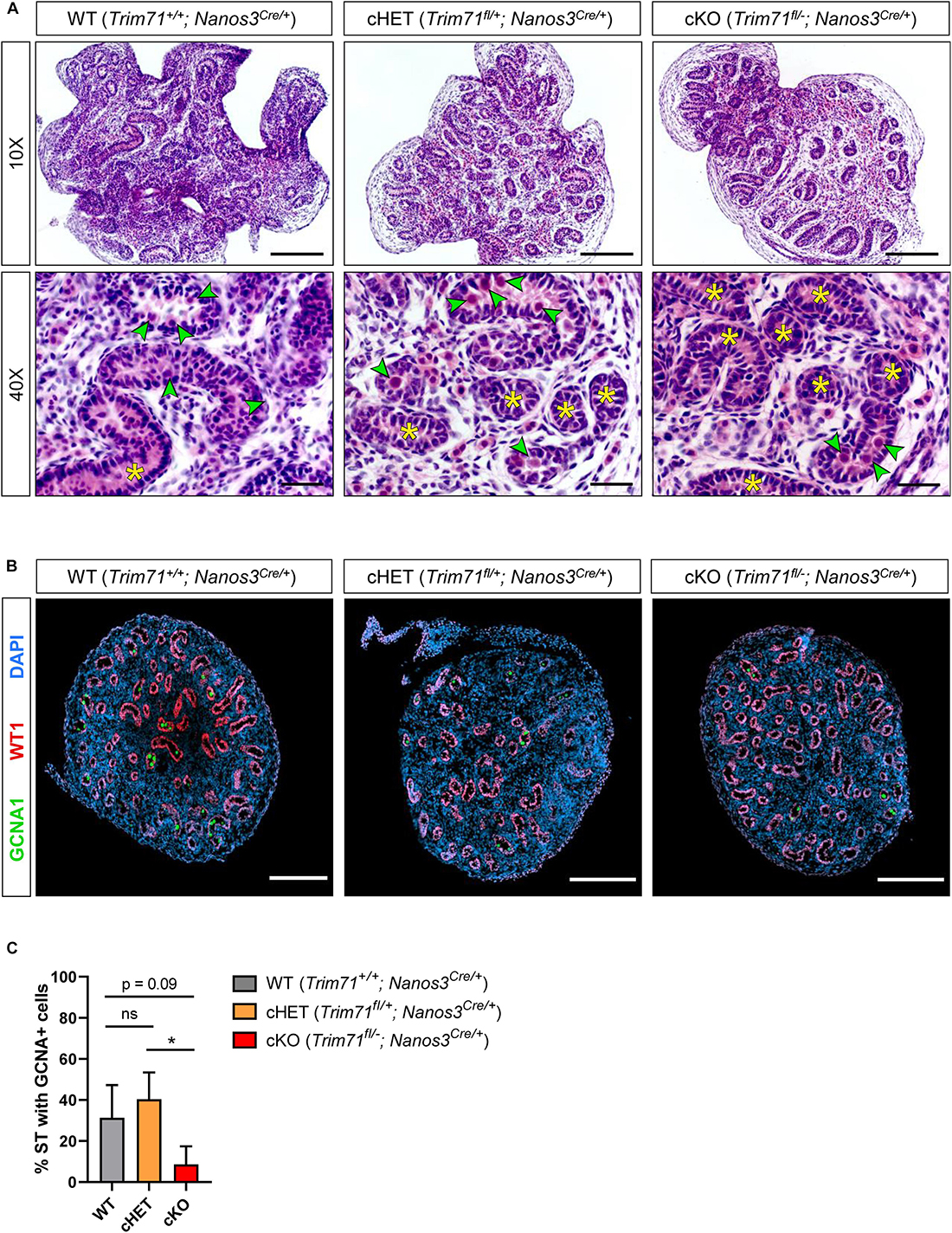
Figure 3. Infertility in germline-specific Trim71 cKO male mice has an embryonic origin. (A) Representative H&E stainings on paraffin testes cross-sections from neonatal (P0.5) wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) male mice. Gonocytes within the seminiferous tubules are marked with a green arrow head, and gonocyte-lacking seminiferous tubules are marked with a yellow asterisk (*) in 40x magnification images. Scale bars represent 100 and 20 μm in 10× and 40× magnifications, respectively. (B) Representative immunofluorescence stainings on testes cryo-sections from neonatal (P0.5) wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/−; Nanos3Cre/+) male mice. Images show co-staining with GCNA1 (gonocytes), WT1 (Sertoli cells) and DAPI (nuclei). Scale bars represent 200 μm. (C) Quantification of seminiferous tubules (ST) containing GCNA+ cells per testis cross-section from B, depicted as percentages. Error bars represent SD (n = 3). *P-value < 0.05, ns, non-significant (unpaired Student’s t-test). See also Supplementary Figures 4–6.
In vitro PGC Specification Reveals Reduced Numbers of PGCLCs Derived From Trim71-Deficient ESCs
In order to determine whether Trim71 deficiency causes PGC specification defects, wild type (WT, Trim71fl/fl) and Trim71 knockout (KO, Trim71–/–) murine ESCs (Mitschka et al., 2015) were differentiated into PGC-like cells (PGCLCs) in vitro as previously described (Hayashi et al., 2011). In short, ESCs (d0) growing under naïve conditions were primed to epiblast-like cells (EpiLCs) (d2) for two days in monolayer culture and differentiated into PGCLCs (d8) for six more days in the presence of BMP4 while growing as spheroids (Supplementary Figure 5A). ESCs (d0) represent in vivo embryonic stem cells of the blastocyst’s inner cell mass at E3.5-4.5 while EpiLCs (d2) represent in vivo epiblast stem cells (EpiSCs) at E5.5-6.5, and PGCLCs (d8) correspond to in vivo PGCs at E9.5 (Hayashi et al., 2011). Thus, several markers were measured in bulk cell populations at d0, d2 and d8 to monitor the differentiation process (Supplementary Figure 6). Of note, these experiments are rather sensitive to small variations in experimental conditions and need to be interpreted with caution.
Both WT and Trim71 KO ESCs were primed to EpiLCs as shown by a decrease of the naïve pluripotency marker Klf4 (Guo et al., 2009; Hayashi et al., 2011) and a simultaneous increase of the primed pluripotency epiblast-specific marker Dnmt3b (Hayashi et al., 2011; Veillard et al., 2014) at d2 (Supplementary Figures 6A,B). The subsequent Dnmt3b downregulation observed at d8 was indicative of a successful specification induction by BMP4, as the early PGC marker Prdm14 represses Dnmt3b to enable PGC epigenetic reprogramming (Grabole et al., 2013; Seki, 2018). Accordingly, both WT and Trim71 KO cells showed a significant upregulation of PGC-specific markers at d8 compared to d2 including Prdm14, Prdm1/Blimp1 and their downstream targets Tfap2c and Nanos3 (Supplementary Figures 6C–F). However, the levels of Blimp1 at d8 were significantly lower in Trim71 KO cells than in WT cells, an effect that was also apparent – although not significant – for Nanos3. Importantly, while an upregulation of the early PGC marker Nanos3 (Tsuda et al., 2003) was already observable at d8, the expression of the late PGC marker Ddx4/Vasa (Fujiwara et al., 1994; Tanaka et al., 2000) was not increased (Supplementary Figure 6G), indicating that the in vitro-generated PGCLCs represent in vivo PGCs prior to the time point of Ddx4 expression (E10.5), as expected (Hayashi et al., 2011).
In order to estimate the number of PGCLCs specified from WT and Trim71 KO ESCs, d8-bulk populations were stained for the surface markers ITGB3/CD61 (PE-Cy7) and SSEA-1/CD15 (APC) – previously reported to unequivocally identify PGCLCs (Hayashi et al., 2011) – and were analyzed by flow cytometry. We found that the number of PGCLCs derived from Trim71-deficient ESCs was reduced by about 25% (Supplementary Figures 5B,C). A significant decrease of the PGC-specific marker SSEA-1/CD15 (APC) – but not of ITGB3/CD61 (PE-Cy7) – was also observed in PGCLCs derived from Trim71-deficient ESCs (Supplementary Figures 5D,E). However, these stainings do not show the expected distinct PGCLC population (Hayashi et al., 2011), which might be indicative of an incomplete induction.
Of note, Oct4 is required for the maintenance of PGCs after their specification (Kehler et al., 2004). The highest expression of Oct4 in themouse embryo is observed in the early blastocyst and decreases progressively in the epiblast upon gastrulation until being restricted to PGCs at E7.5 (Rosner et al., 1990; Ecsedi and Grosshans, 2013). Accordingly, we detected a high Oct4 expression in ESCs (d0) which significantly decreased upon priming to EpiLCs (d2) and was maintained in PGCLCs (d8) upon specification (Supplementary Figure 6H). A similar pattern was observed for Trim71 expression in WT cells (Supplementary Figure 6I) with a decrease in its mRNA level after priming (d2) and a sustained expression in PGCLCs (d8). While our results have revealed a moderate reduction in the number of Trim71-deficient PGCLCs upon induction, we believe that this cannot fully explain the dramatic phenotype observed in Trim71 cKO mice. This, together with the sustained Trim71 expression in PGCLCs at d8, suggests a further role for TRIM71 downstream of PGC specification.
TRIM71 Deficiency Does Not Impair in vivo Migration of PGCs Toward the Genital Ridges
After specification, PGCs migrate along the hindgut toward the genital ridges (Tam and Snow, 1981; Anderson et al., 2000). Thus, we next investigated whether TRIM71 is required for PGC migration in vivo. To this end, we used full Trim71 KO (Trim71–/–) embryos resulting from the mating of full heterozygous (Trim71fl/–) animals in order to exclude an incomplete Trim71 deletion at this stage resulting from an insufficient Nanos3-driven Cre expression. Wild type (Trim71fl/fl) and full KO (Trim71–/–) embryos at stages E8.5-8.75 were dissected to isolate the hindgut and alkaline phosphatase (AP)-positive PGCs were stained. PGCs were detected in close proximity to the allantois in both wild type and Trim71-deficient embryos (Supplementary Figure 7). Although our AP-stainings do not provide quantitative information, they clearly show that Trim71 expression is not essential for migration of PGCs toward the genital ridges and furthermore reinforce the notion that no major PGC specification defects are apparent, as already suggested by our PGCLC induction experiments. Thus, TRIM71 likely affects germ cell development at later embryonic stages.
TRIM71 Controls Proliferation in the PGC-Like in vitro Model Cell Line TCam-2
After arriving at the genital ridges, the PGC founder population strongly increases in numbers before undergoing sex determination (Tam and Snow, 1981; Anderson et al., 2000; Bowles and Koopman, 2010; Ewen and Koopman, 2010). Since TRIM71 is known to regulate proliferation in several cell lines (Chen et al., 2013; Ren et al., 2018; Hu et al., 2019; Torres-Fernández et al., 2019), we next wanted to investigate whether TRIM71 may control proliferation also in PGCs. Because the isolation of sufficient PGCs from genital ridges for its absolute reliable quantification is technically very challenging, we used the human GCT-derived seminoma cell line TCam-2, which is a widely accepted in vitro PGC model (Irie et al., 2015; Sugawa et al., 2015; Nettersheim et al., 2016a, b, c; Mitsunaga et al., 2017; Müller et al., 2020) and expresses high TRIM71 levels, similar to other GCT-derived cell lines (Supplementary Figure 8A). To this end, we knocked down (KD) TRIM71 with two different siRNAs (Figure 4A), and monitored the increase in cell numbers over time (Figure 4B). Indeed, TRIM71 KD cells proliferated significantly slower in the course of the experiment, as shown by area under curve (AUC) measurements (Figure 4C). Consistent with our previous observations in PGCLCs, TRIM71 KD TCam-2 cells showed a significant downregulation of BLIMP1 and NANOS3, while TFAP2C and DDX4 remained unaltered (Figures 4D–G). Although our data do not provide evidence for a reduction in PGC proliferation in vivo, these results suggest that TRIM71 may be important for the expansion of the germ cell pool during embryogenesis, which could be an explanation for the reduction of male gonocytes observed in neonatal Trim71 cKO mice.
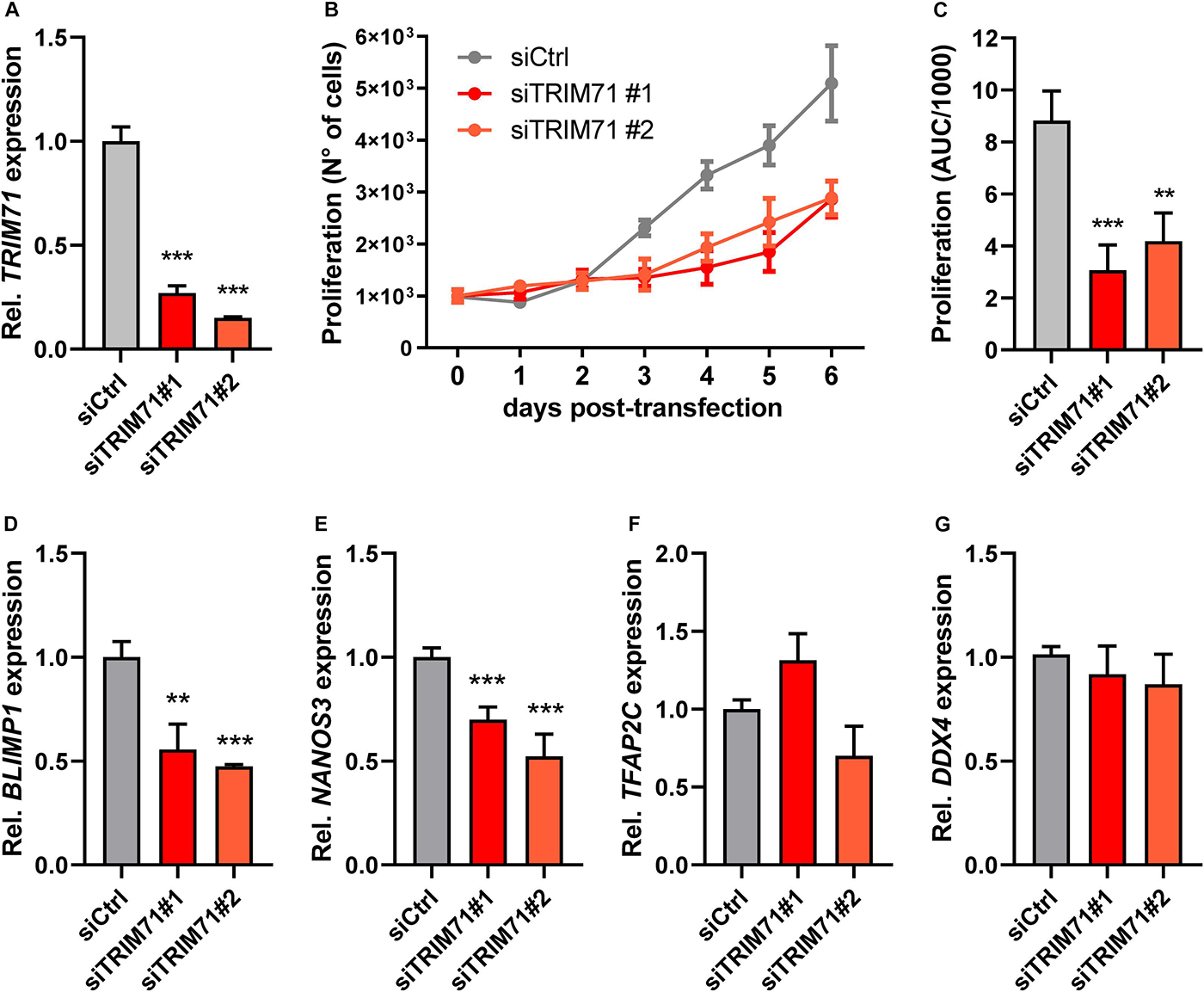
Figure 4. TRIM71 controls proliferation in the in vitro PGC-like model cell line TCam-2. (A) RT-qPCR measurement of TRIM71 in wild type (siCtrl) and TRIM71 KD (siTRIM71 #1 and siTRIM71 #2) TCam-2 cells 72 h post-transfection (n = 3). (B) Proliferation of wild type (siCtrl) and TRIM71 KD (siTRIM71 #1 and siTRIM71 #2) TCam-2 cells depicting the increase in cell numbers over time after transfecting them with siRNAs and seeding them in equal numbers (d0) (n = 5). (C) Area under curve (AUC) measurements for the proliferation curves depicted in B (n = 5). (D–G) RT-qPCR measurements for the indicated PGC markers in wild type (siCtrl) and TRIM71 knockdown (siTRIM71 #1 and siTRIM71 #2) TCam-2 cells 72 h post-transfection (n = 3–6). For all RT-qPCRs, the housekeeping gene 18S rRNA was used for normalization. For all graphs, error bars represent the SEM. ***P-value < 0.005, **P-value < 0.01 (unpaired Student’s t-test). See also Supplementary Figure 8.
TRIM71 Controls Proliferation in the Germline-Derived Tumor Cell Line NCCIT
The failure of male PGCs to further differentiate into gametes can lead to the development of testicular GCT (TGCT) (Raman et al., 2005; Looijenga et al., 2010; Zorrilla and Yatsenko, 2013). TGCT can be divided into three groups: (I) tumors of newborns and infants (teratomas and yolk sac tumors), (II) tumors of adolescents and young adults (seminomas and non-seminomas) and (III) spermatocytic seminoma of elderly men (≥ 50 years) (Oosterhuis and Looijenga, 2005). TRIM71 is highly expressed in several GCT-derived cell lines (Rybak et al., 2009; Chang et al., 2012; Torres-Fernández et al., 2019) (Supplementary Figure 8A), and is upregulated in TGCT seminoma and non-seminoma patients (Tang et al., 2017; Supplementary Figures 8B–D). Since we have shown that TRIM71 controls proliferation of seminoma TCam-2 cells, we evaluated whether TRIM71 could also control proliferation in the GCT-derived non-seminoma cell line NCCIT. Because TRIM71 is strongly expressed in NCCIT cells (Supplementary Figure 8A) and we did not achieve a satisfying TRIM71 depletion via KD (data not shown), we generated TRIM71 frameshift mutations via CRISPR/Cas9. To this end, we used two different single guide RNAs (sgRNA), one targeting the N-terminal RING domain and another targeting the C-terminal NHL domain (Supplementary Figure 8E). For the RING sgRNA (ΔRING), the generation of single NCCIT clones strikingly showed that an 83 kDa N-truncated RINGless version of TRIM71 is generated from the use of an alternative in-frame ATG codon present downstream of the targeting region (Supplementary Figure 8F). For the NHL sgRNA (ΔNHL6), generation of single clones showed that the resultant protein was either unstable or had a C-terminal truncation of the last NHL repeat (Supplementary Figure 8G), a mutation which is already known to mimic the full KO phenotype in vivo (Schulman et al., 2008) and to impair mRNA binding and repression in vitro (Torres-Fernández et al., 2019).
Single cell clones often have a selection bias toward robust in vitro proliferation and survival. In order to study TRIM71-dependent cell proliferation in an unbiased manner, we analyzed cell population dynamics during in vitro growth competition assays using non-clonal mixed pools of wild type and TRIM71 mutant cells. To this end, NCCIT cells were mock-transfected with an empty vector (EV) or with TRIM71-targeting vectors (ΔRING or ΔNHL6) and sorted for Cas9-GFP + cells. Bulk-transfected EV (wild type) and ΔRING/ΔNHL6 (TRIM71 mutant) NCCIT pure populations (Supplementary Figures 9A,B) were mixed in a 1:1 ratio. We then evaluated changes in the distribution of single reads via NGS (Illumina MiSeq system) (Ravi et al., 2018) for wild type and TRIM71 mutant alleles over a time period of 21 days and classified them as wild type reads, reads with TRIM71 frameshift (loss-of-function) mutations and reads with TRIM71 in-frame mutations (Figure 5). As a negative control, pure wild type NCCIT (EV) cell populations were analyzed at day 0 and day 21, showing no relevant changes in allele distribution (Supplementary Figures 9C,D). In contrast, the percentages of alleles with TRIM71 frameshift mutations dropped ~2-fold (from 11.34% to 5.96%) for NCCIT ΔRING cells within 21 days (Figure 5A) and ~13-fold (from 32.46% to 2.53%) for NCCIT ΔNHL6 cells in the same time period (Figure 5B). Interestingly, TRIM71 in-frame mutations also resulted in a growth disadvantage, although to a lesser degree in each case (Figures 5A,B).
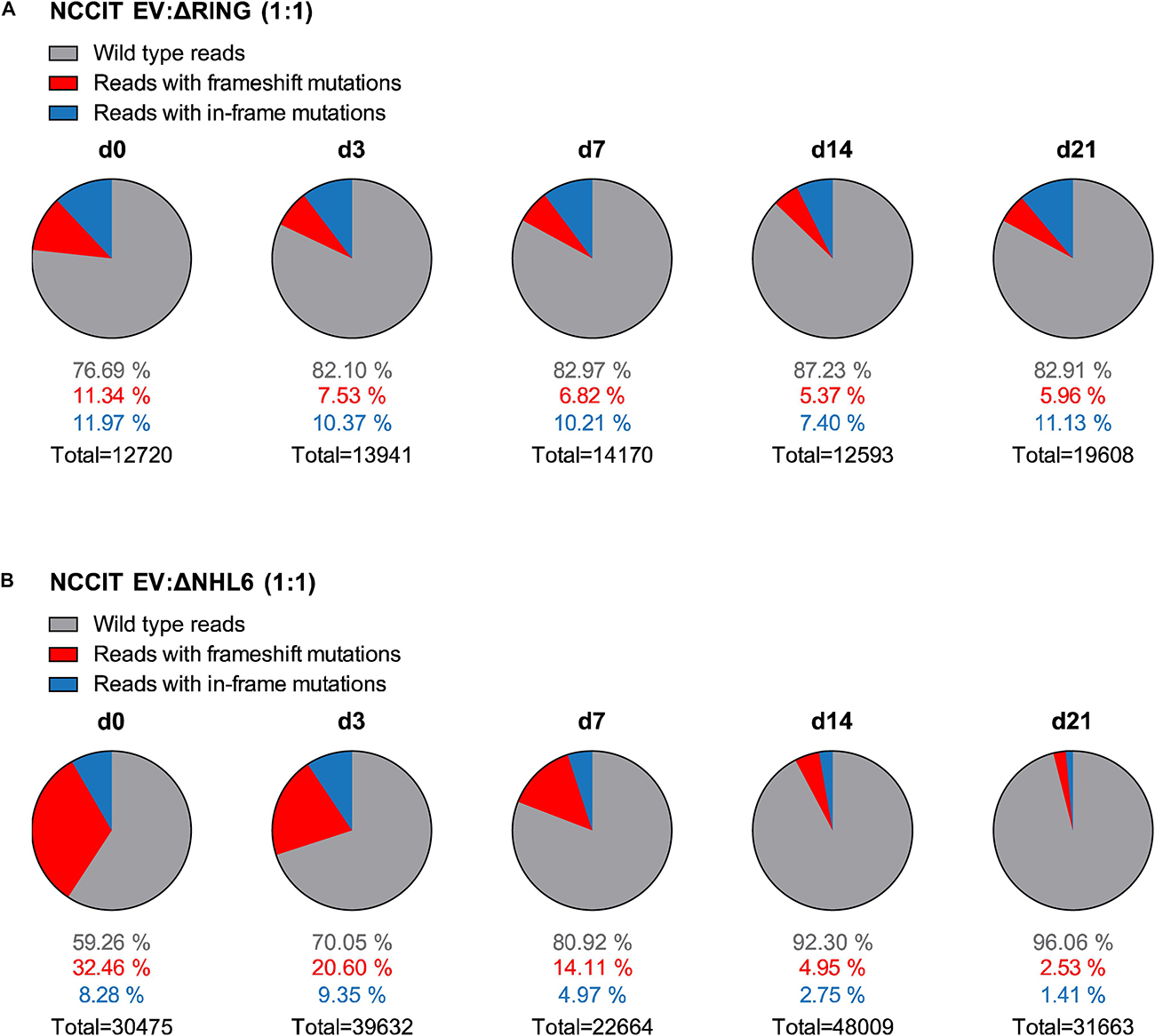
Figure 5. NCCIT cells with TRIM71 mutations show population maintenance defects in growth competition assays. (A) Pie charts showing allele frequencies for wild type (EV) and TRIM71 RING mutant (ΔRING) or (B) TRIM71 NHL mutant (ΔNHL6) NCCIT cells obtained at different time points during growth competition assays. For the assay, NCCIT wild type cells (EV) were mixed 1:1 with either TRIM71 RING mutant (ΔRING) or TRIM71 NHL mutant (ΔNHL6) NCCIT cells and directly analyzed (d0) before their culturing, following by subsequent analysis at several time points (d3, d7, d14, and d21). Allele frequency was analyzed via NGS using the Illumina MiSeq platform. For TRIM71 mutant alleles, reads with in-frame mutations and frameshift mutations (loss-of-function) in each respective domain (RING or NHL) are depicted. The total number of sequencing reads for each time point is indicated under each respective pie chart. See also Supplementary Figures 8–10.
In the course of these growth competition assays with NCCIT cells, we neither observed morphological changes indicative of differentiation nor signs of abnormal cell death upon regular media change, suggesting that the observed changes in population dynamics may derive from TRIM71-mediated control of proliferation. We then conducted proliferation assays on individual ΔRING and ΔNHL6 NCCIT clones for confirmation. This time, we employed the eFluor670 proliferation dye to stain NCCIT cells (d0) and measured the loss of fluorescence intensity over time as a readout for proliferation. While eFluor670 (APC) staining was comparable for wild type (EV Ctrl) and TRIM71 mutant (ΔRING or ΔNHL6) NCCIT cells at the beginning of the experiment (d0), a retardation of eFluor670 (APC) histogram could be observed for both TRIM71 ΔRING and ΔNHL6 NCCIT cells at the end of the experiment (d4), revealing proliferation defects in TRIM71 mutant NCCIT cells (Supplementary Figures 10A,B). Based on the fluorescence intensity of the eFluor670 dye at d0 and d4, and assuming that fluorescence intensity halves upon every cell division, the number of cell divisions as well as the average duration of the cell cycle can be estimated (Supplementary Figures 10C,D). The cell cycles of ΔRING and ΔNHL6 mutants were 1.68 h and 3.94 h longer, respectively, than that of wild type NCCIT cells. Such a prolongation of the cell cycle corresponds to an 11.2% and 25.2% reduction in proliferation for ΔRING and ΔNHL6 mutants, respectively. The stronger effect of the ΔNHL6 mutant on proliferation may be due to the regulation of the cell cycle inhibitor CDKN1A (Supplementary Figure 10E), which is known to be dependent on TRIM71’s NHL domain and independent of its RING domain (Torres-Fernández et al., 2019). These results collectively demonstrate that TRIM71 is involved in the control of proliferation in NCCIT cells, and are consistent with the data obtained with TCam-2 cells as well as with previous studies involving other tumor cell lines (Chen et al., 2013; Ren et al., 2018; Hu et al., 2019; Torres-Fernández et al., 2019). Our work thereby confirms a role for TRIM71 supporting the proliferation of GCT-derived cells. Along with the elevated TRIM71 expression observed in TGCT patients, these findings suggest that TRIM71 may not only affect germ cell proliferation during developmental processes, but may also contribute to the malignancy of GCT.
We then measured the expression of the PGC-specific markers NANOS3 and BLIMP1 in NCCIT wild type and TRIM71 mutant cells. In line with our previous findings in Trim71 KO PGCLCs and TRIM71 KD TCam-2 cells, NANOS3 was downregulated in TRIM71 ΔNHL6 mutant NCCIT cells (Supplementary Figure 10F) and BLIMP1 was downregulated in both ΔRING and ΔNHL6 mutants (Supplementary Figure 10G). Interestingly, we and others recently found that TRIM71 is not only an mRNA repressor, but is also capable of positive mRNA regulation via direct binding and stabilization (Foster et al., 2020; Torres-Fernández et al., 2021). Thus, we checked several published datasets for Blimp1 and Nanos3 expression and mRNA binding. In accordance with our data, Blimp1 was found consistently downregulated in Trim71 KO, RING mutant and NHL mutant ESCs and its mRNA was co-precipitated with TRIM71 in RNA-IPs (Supplementary Figure 10H; Welte et al., 2019) while Nanos3 mRNA was not (data not shown). Similarly, BLIMP1 was downregulated in TRIM71 KO hepatocellular carcinoma (HCC) cells (Supplementary Figure 10I; Welte et al., 2019), and BLIMP1 mRNA was also co-precipitated with TRIM71 in HCC cells (Supplementary Figure 10I; Foster et al., 2020). These results suggested a post-transcriptional regulation of Blimp1 mRNA mediated by TRIM71, which is known to directly interact with mRNAs via conserved RNA hairpins (Kumari et al., 2018; Torres-Fernández et al., 2019; Welte et al., 2019). Supporting the RNA-IP binding data, several TRIM71-binding hairpins were found located along the Blimp1 mRNA (Supplementary Figure 10J) while no hairpins were identified along the Nanos3 mRNA (Welte et al., 2019). The ability of TRIM71 to positively regulate directly bound mRNAs has been reported to depend on both the RING and NHL domains, and was achieved via TRIM71-mediated repression of let-7 miRNA (Torres-Fernández et al., 2021). Interestingly, this mechanism was observed in both ESCs and HCC cells (Torres-Fernández et al., 2021). Thus, it is possible that TRIM71 regulates Blimp1 expression via this mechanism, since (i) Blimp1 is a known let-7 target (West et al., 2009), (ii) TRIM71 is able to directly bind Blimp1 mRNA and (iii) TRIM71-mediated Blimp1 regulation depends on both the RING and NHL domains.
Exome Sequencing Data Identifies TRIM71 Variants in Infertile Men With Severely Impaired Spermatogenesis
The causes of the SCO phenotype in men remain poorly understood (Tüttelmann et al., 2011; Tüttelmann et al., 2018; Koc et al., 2019). In order to identify novel associated genes, we utilized exome sequencing data of 247 SCO subjects belonging to the Male Reproductive Genomics (MERGE) study. We employed our in-house software Sciobase and our newly developed software Haystack to analyze loss-of-function (LoF) variants. After strict filtering based on quality criteria, minor allele frequency (MAF) in the general population, high expression in human testes (GTEx) and absence of LoF variants in individuals with complete spermatogenesis, we identified a total of 721 genes with LoF variants specifically found in SCO patients (Figures 6A–C). Via this independent and unbiased approach, we found TRIM71 to be present among these genes, revealing a possible association with human male infertility. This finding is strongly supported by our previous data showing that TRIM71 deficiency results in an SCO-like phenotype in male mice. Of note, most of the identified human genes carried LoF variants in only one patient within the studied cohort and none of the identified genes carried LoF variants in more than four individuals, indicating that the SCO phenotype is highly heterogeneous (Figure 6B). Similar results were observed upon inclusion of genes with lower expression in the testis (Figure 6C).
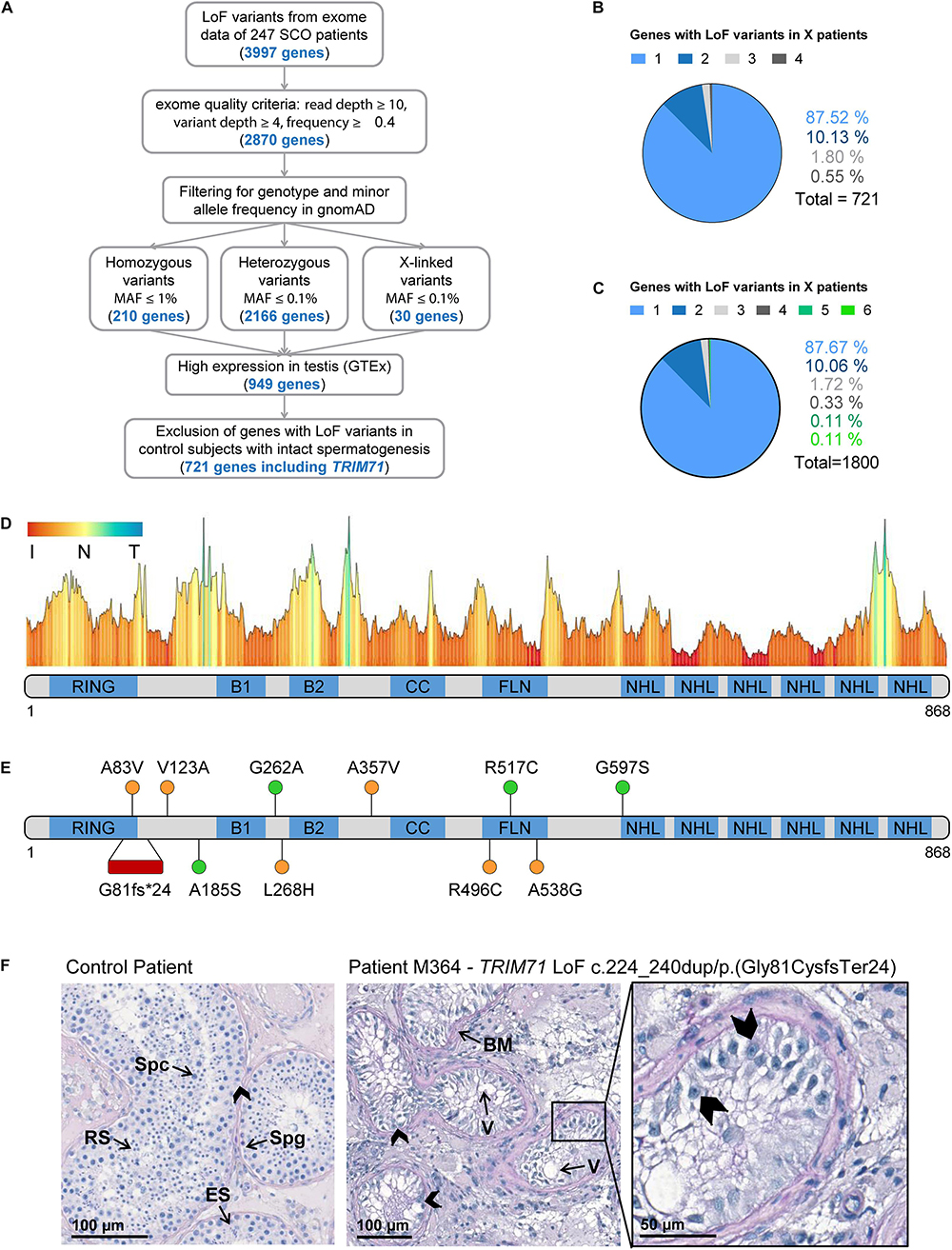
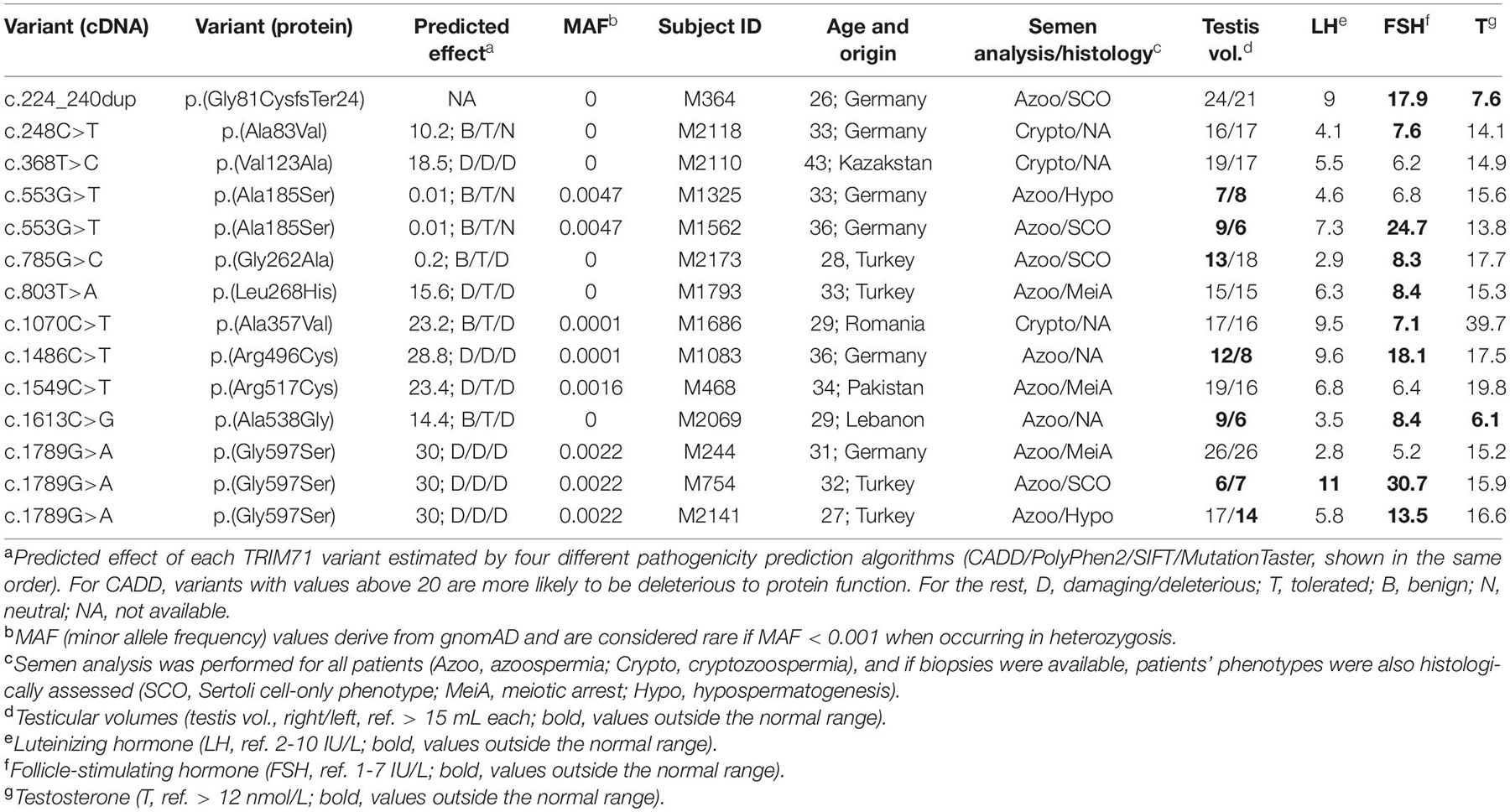
Figure 6. Exome sequencing in infertile SCO patients reveals an association of TRIM71 deficiency with human male infertility. (A) Filtering scheme detailing gene prioritization of exome sequencing data from 247 individuals with Sertoli cell-only (SCO) phenotype using Sciobase and Haystack (see details in the section “METHODS”). Gene numbers remaining after each filtering step are indicated in blue. Loss-of-function (LoF) variants include stop, frameshift, splice acceptor and splice donor variants. gnomAD, Genome Aggregation Database; MAF, minor allele frequency; GTEx, Genotype-Tissue Expression Portal. (B) Percentages of genes (total = 721, obtained at the end of the filtering process depicted in A) affected by LoF variants in 1–4 SCO patients. None of the genes carried LoF variants in more than 4 patients in the SCO cohort (n = 247). (C) Percentages of genes (total = 1800, obtained at the end of the filtering process depicted in A, excluding “High expression in testis - GTEx” filtering step) affected by LoF variants in 1-6 SCO patients. None of the genes carried LoF variants in more than 6 patients in the SCO cohort (n = 247). (D) TRIM71 mutation tolerance landscape obtained from MetaDome (https://stuart.radboudumc.nl/metadome/). The graph displays missense over synonymous variant ratios per position for the entire protein, based on gnomAD variants. I, intolerant; N, neutral; T, tolerant. (E) Schematic representation of TRIM71 domain structural organization depicting the location of the genetic variants identified in MERGE. Red, presumably pathogenic LoF variants; orange, missense variants of uncertain significance; green, missense variants found also in proven fathers. (F) Periodic acid–Schiff stainings of testis sections obtained by testicular biopsy from a control patient and an SCO patient (subject M364), who harbors the TRIM71 LoF variant c.224_240dup/p.(Gly81CysfsTer24) in heterozygosis. The control section shows intact spermatogenesis (Spg, spermatogonia; Spc, spermatocytes; RS, round spermatids; ES, elongated spermatids) as opposed to subject M364, whose histological evaluation revealed an SCO phenotype (exemplary Sertoli cells indicated by arrow heads) with several degenerated seminiferous tubules, frequently including large vacuoles (V) and thickened basement membranes (BM). See also Table 1, Supplementary Figures 11, 12, and Supplementary Tables 1, 2.
For a further evaluation of TRIM71 as a candidate gene associated with human male infertility, we screened 908 infertile patients - including those with SCO from MERGE - for variants in TRIM71. We identified 11 different rare (MAF ≤ 0.001) heterozygous TRIM71 variants, including the aforementioned LoF variant and 10 missense variants, present in a total of 14 infertile individuals with severely impaired spermatogenesis and varying histological phenotypes (Figures 6D–F and Table 1). The majority of subjects (11 out of 14) were azoospermic, and some of them presented with reduced testicular volumes, elevated luteinizing hormone (LH) levels, elevated follicle-stimulating hormone (FSH) levels and/or reduced serum testosterone (T) levels (Table 1), all signs of broad testicular dysfunction present in 60% of azoospermic men (Tüttelmann et al., 2018).
Generally, although most of the identified missense variants were classified as variants of uncertain significance according to ACMG/AMP guidelines (Supplementary Table 1), TRIM71 seems to be rather intolerant to missense variation (Figure 6D; Wiel et al., 2019). This was also indicated by a high Z score value of 3.28 for TRIM71. The Z score is a metric computed by gnomAD database and ranges from −5 to 5, with higher values indicating an intolerance to variation and, therefore, a higher likelihood for TRIM71 variants to disrupt TRIM71 function. Furthermore, parameters such as MAF or pathogenicity prediction algorithms were used to estimate the reliability for the association of each variant with male infertility (Table 1). The conservation of the affected residues for each variant was also evaluated (Supplementary Figure 11A). Four of the TRIM71 missense variants identified in infertile men were also found in control subjects with complete spermatogenesis (n = 89) or in a Dutch cohort of proven fathers (n = 5784), making their association with male infertility rather unlikely (Figure 6E, green). In contrast, other identified TRIM71 missense variants were present in one patient each but neither in control subjects nor in the Dutch cohort of proven fathers (Figure 6E, orange). Of those, c.368T>C/p.(Val123Ala), c.803T>A/p.(Leu268His), c.1070C>T/p.(Ala357Val) and c.1486C>T/p.(Arg496Cys) were considered damaging by at least two out of four pathogenicity prediction algorithms (Table 1), and the respective residues for those variants were highly conserved among vertebrates (Supplementary Figure 11A). Thus, these variants have a higher likelihood of being associated with male infertility.
As an additional line of evidence, all patients carrying variants in TRIM71 (Table 1) were evaluated for relevant variants in other genes previously reported in association with male infertility (n = 181, listed in Supplementary Table 2). Importantly, one heterozygous frameshift variant was found for the gene SYCP2 in patient M1686, who carries the TRIM71 variant c.1070C>T/p.(Ala357Val). SYCP2 was recently associated with male infertility (Schilit et al., 2019), and it is thus unlikely that this patient’s TRIM71 variant alone is responsible for his cryptozoospermia, but an oligogenic cause of his phenotype cannot be ruled out. Furthermore, a heterozygous missense variant for the gene NNT was identified in patient M1083, who carries the TRIM71 variant c.1486C>T/p.(Arg496Cys). In this case, the NNT missense variant is of unclear significance according to ACMG/AMP guidelines, and its functional contribution to the patients’ phenotype can therefore neither be confirmed nor excluded at this point. Collectively, our analysis leaves the TRIM71 variants c.368T>C/p.(Val123Ala) and c.803T>A/p.(Leu268His) as the likeliest candidates for being associated with male infertility.
Exome Sequencing Data Reveals an Association of TRIM71 LoF Variants With the SCO Phenotype
As mentioned above, a TRIM71 LoF variant - c.224_240dup/p.(Gly81CysfsTer24) - was identified via exome sequencing in subject M364, who presented with a clear SCO phenotype (Figures 6E,F and Table 1). This variant consists of a duplication of 17 nucleotides which generates a frameshift resulting in a premature termination codon (PTC) downstream of the RING domain (Supplementary Figure 11B). This particular TRIM71 LoF variant has not been described in any public database so far and was classified as likely pathogenic according to ACMG/AMP guidelines (Supplementary Table 1). Furthermore, the observed/expected (o/e) score computed for TRIM71 in the gnomAD database was extremely low (o/e score = 0.04, 90% confidence interval 0.01–0.17). The o/e score compares the number of observed versus theoretically expected LoF variants for a gene of interest. Without any selection pressure applied, an o/e ratio of around 1 would be expected for any given gene, whereas values below 0.35 are a rather clear sign of selection pressure against LoF variants, likely leading to haploinsufficiency intolerance. Furthermore, subject M364 carries neither additional LoF variants nor likely pathogenic missense variants in any other known infertility-associated genes (listed in Supplementary Table 2). This patient presented with normal testicular volumes and LH levels, but increased FSH and decreased testosterone levels (Table 1), as is often the case in men with impaired spermatogenesis. Of note, no TRIM71 LoF variants were found in the Dutch cohort of proven fathers, and only one LoF variant was present in the exome data from 125,748 presumably healthy individuals from gnomAD (Karczewski et al., 2020). This constitutes a significant enrichment of TRIM71 LoF variants in the MERGE cohort (n = 908; p = 0.01) and an even higher enrichment in the SCO subcohort (n = 247; p = 0.004), supporting a reliable association of TRIM71 LoF variants with human male infertility. Thus, the reported TRIM71 LoF variant is likely the cause of subject M364’s SCO phenotype, underscoring a haploinsufficiency of TRIM71 in human spermatogenesis. In contrast, although sperm counts were strongly reduced in TRIM71 heterozygous mice, these animals were fertile. This discrepancy may be attributed to inherent mechanistic differences between murine and human gametogenesis. Indeed, it has been reported for multiple diseases that the modulation of a specific gene can result in different severity and range of symptoms for mice and humans (Elsea and Lucas, 2002).
In order to address the functionality of all identified TRIM71 variants, we overexpressed them in HEK293T cells to evaluate protein stability via western blot analysis. Whereas all TRIM71 missense variants yield stable proteins, the LoF variant resulted in a functional KO as expected, since no protein was detected via western blot analysis for the respective LoF construct Flag-dupPTC (Supplementary Figure 12A). We additionally tested the ability of the different TRIM71 constructs to repress the 3′UTR of CDKN1A (Torres-Fernández et al., 2019) as well as the activity of let-7 miRNA (Torres-Fernández et al., 2021) via luciferase reporter assays in HEK293T cells. CDKN1A 3′UTR repression was impaired for the Flag-dupPTC construct, and significantly diminished for the G597S variant which affects TRIM71’s NHL domain (Supplementary Figures 12B,C). The ability of TRIM71 to derepress a luciferase reporter under the control of a 3′UTR with multiple let-7 binding sites was also abrogated for the Flag-dupPTC construct (Supplementary Figures 12D,E). These assays collectively show that the TRIM71 LoF variant identified in the patient with SCO phenotype yields a functional KO. Future experiments should determine whether the here identified missense variants impair other known TRIM71 functions (e.g., TRIM71’s E3 ubiquitin ligase role) as well as determine their pathogenicity in the context of human infertility.
Discussion
TRIM71 is a stem cell-specific protein expressed early in development and with an essential function for embryogenesis (Schulman et al., 2005, 2008; Rybak et al., 2009; Cuevas et al., 2015). Embryonic lethality in Trim71 full KO mice is accompanied by neural tube closure defects, revealing also a function of TRIM71 in the development of the nervous system (Schulman et al., 2005, 2008; Rybak et al., 2009; Cuevas et al., 2015). Indeed, TRIM71 is known to promote self-renewal of neural progenitor cells (Chen et al., 2012), and TRIM71 variants affecting its NHL domain have been associated with human congenital hydrocephalus, a brain developmental disease characterized by enlarged brain ventricles due to an abnormal accumulation of cerebrospinal fluid (Furey et al., 2018). Furthermore, TRIM71 has been connected with carcinogenesis in several studies (Chen et al., 2013, 2019; Ren et al., 2018; Hu et al., 2019; Torres-Fernández et al., 2019). Altogether, previous research on TRIM71 has been mostly focused on embryonic and neural stem cells as well as cancer cells. Our work here reports a novel role for TRIM71 in the development of germ cells during embryogenesis with crucial implications in murine and possibly human fertility.
In line with previous studies (Rybak et al., 2009; Du et al., 2020), our work reveals an expression of Trim71 in adult mouse testes which we found to be confined to SSCs in mice and human. We showed that germline-specific ablation of Trim71 early in mouse development causes a substantial reduction of gonad size and infertility in both sexes. Further characterization of adult Trim71 cKO mouse testes revealed a significant downregulation of SSC-specific and spermatid markers caused by a strong reduction in the number of developing germ cells, a condition which in humans is known as Sertoli cell-only (SCO) phenotype (Tüttelmann et al., 2011, 2018; Koc et al., 2019). Via exome sequencing of human infertile men, we uncovered an association of TRIM71 deficiency with the SCO phenotype.
In humans, infertility affects 10–15% of couples trying to conceive (Zorrilla and Yatsenko, 2013). Male factors contribute in about half of couples and usually genetic causes correlate with a more severe spermatogenic impairment (Lee et al., 2011). Lately, several genes have been identified as monogenic causes for azoospermia due to meiotic arrest (e.g., TEX11 Yatsenko et al., 2015, STAG3 Van Der Bijl et al., 2019, M1AP Wyrwoll et al., 2020 and SHOC1 Krausz et al., 2020). However, the SCO phenotype seems to be a highly heterogeneous condition, as indicated by our data, making the identification of monogenic causes especially challenging. Our work uncovers TRIM71 as the first monogenic cause for this condition, based on the SCO-like phenotype observed in our germline-specific Trim71 cKO mouse together with the TRIM71 LoF variant that we found in an SCO patient. The finding of TRIM71 as a novel SCO candidate gene is relevant in terms of patient care, as it may allow future patients to be provided with a causal diagnosis for their azoospermia, and the potential success of testicular biopsy and sperm extraction with the aim to perform in vitro fertilization could be predicted in advance.
In contrast to the convincing relevance of the TRIM71 LoF variant, assessing the role of the identified rare TRIM71 missense variants in infertile patients of varying histological phenotypes is much more challenging. Of note, variants in the same gene may cause a spectrum of histological phenotypes (Wyrwoll et al., 2020). Although our analysis provides a prediction for the pathogenicity of these variants, their functional effects on human male fertility are yet of unclear significance and can be only determined experimentally. Nevertheless, it is worth emphasizing that most missense variants identified in TRIM71 cluster outside of the NHL domain, as mutations affecting the NHL domain have been proven to be highly deleterious (Slack et al., 2000; Kumari et al., 2018) and compromise survival during early embryogenesis (Schulman et al., 2008). Accordingly, we found that these missense variants do not compromise NHL-dependent TRIM71 functions. Future experiments using these TRIM71 variants should determine whether they functionally affect TRIM71’s E3 ubiquitin ligase role, and if so, how exactly TRIM71-mediated ubiquitylation contributes to germ cell development.
Our germline-specific Trim71 cKO mouse model provided further evidence for the role of Trim71 in germ cell development. The SCO-like phenotype was already apparent in the testes of neonatal (P0.5) cKO mice, indicating that TRIM71-induced germline defects have an embryonic origin. However, early developmental defects are likely amplified during postnatal mitotic reactivation and pubertal spermatogenesis as described by a recent work showing infertility in a different germline-specific Trim71 knockout (Trim71–/fl; Ddx4Cre/+) mouse model (Du et al., 2020). In contrast to our observations in Trim71fl/–; Nanos3Cre/+ mice, defects in Trim71–/fl; Ddx4Cre/+ mice were only detected in pubertal (P10) and adult (P56) mice, with germ cell numbers not yet altered in neonatal (P1) mice (Du et al., 2020). This discrepancy may result from an earlier Cre recombinase expression in our cKO model, since Nanos3 is expressed during PGC specification (E7.5) (Ginsburg et al., 1990; Lawson et al., 1999; Tsuda et al., 2003), and Ddx4 is expressed after colonization of the genital ridges (E10.5) (Fujiwara et al., 1994; Tanaka et al., 2000). In fact, Ddx4-induced recombination was previously reported to occur even after sex determination as late as E15.0 (Gallardo et al., 2007). This might also explain why Trim71fl/–; Ddx4Cre/+ females are fertile (Prof. Xin Wu, personal communication, June 22, 2020), while we found Trim71fl/–; Nanos3Cre/+ females to be sterile. Notably, studies in C. elegans showed that the nematode homolog of TRIM71, LIN-41, is required for normal oocyte growth and meiotic maturation, with lin-41 depletion causing sterility in females (Spike et al., 2014; Tsukamoto et al., 2017). Further studies are required to characterize the function of TRIM71 in mammalian female gonad development and fertility.
Although we do not exclude sex-specific functions of TRIM71 in the germline, we found both male and female Trim71 cKO mice to be infertile, suggesting that germ cell developmental defects may occur prior to sex determination of PGCs. Alternatively, TRIM71 deficiency may similarly affect male and female germ cells after sex determination, since published datasets revealed similar TRIM71 expression patterns in male and female PGCs during development. Via in vitro differentiation of ESCs into PGCLCs, we observed reduced numbers of Trim71-deficient PGCLCs at the end of the specification process. Although such a reduction in the PGCLC numbers would underscore mild PGC specification defects of Trim71-deficient cells, it could also result from a specific downregulation of the PGC marker SSEA-1/CD15 which was used for PGCLC staining, and which was found downregulated in Trim71 KO PGCLCs. Furthermore, Trim71-deficient PGCLCs showed a downregulation of Blimp1 and Nanos3, which are two well-known master regulators of PGC development (Kurimoto et al., 2008; Suzuki et al., 2008). Consistently, we also found reduced levels of BLIMP1 and NANOS3 in TRIM71 KD TCam-2 cells and TRIM71 mutant NCCIT cells. We furthermore presented evidence suggesting that TRIM71-mediated BLIMP1 regulation may be attributed to a recently reported novel role for TRIM71 in the repression of let-7 miRNA activity (Torres-Fernández et al., 2021). In this context, it is worth mentioning that the let-7-mediated regulation of Blimp1 plays an important role in murine PGC development (West et al., 2009). Interestingly, Trim71-deficient ESCs showed a premature upregulation of differentiation-promoting miRNAs, including let-7 and several gonad-specific miRNAs (Mitschka et al., 2015; Torres-Fernández et al., 2021), highlighting a yet-unknown role for TRIM71-mediated miRNA regulation in early gonad development. It is however intriguing that TRIM71-mediated Blimp1 regulation does not result in PGC specification or migration defects, as BLIMP1 is well known to control early PGC functions (Ohinata et al., 2005). At this point, we can only speculate that such a moderate regulation of Blimp1 at early stages may induce, for instance, epigenetic changes that may affect later stages of germ cell development. Alternatively, TRIM71-mediated Blimp1 regulation may simply be more relevant at later stages, as the expression of both TRIM71 and BLIMP1 is indeed sustained in developing germ cells even after sex determination (Seisenberger et al., 2012; Zhao et al., 2018).
Since Trim71 expression was still remarkably high in PGCLCs, we aimed at investigating further roles of TRIM71 downstream of PGC specification. While we excluded in vivo PGC migration defects upon TRIM71 depletion, in vitro proliferation assays with TCam-2 and NCCIT cells showed that TRIM71 actively promotes proliferation of these GCT-derived cell lines. Given the similarities of TCam-2 cells – and to a lesser extent of NCCIT cells – with PGCs (Irie et al., 2015; Sugawa et al., 2015; Nettersheim et al., 2016a, b, c; Mitsunaga et al., 2017; Müller et al., 2020), our results suggest that TRIM71 may be involved in the control of PGC proliferation after they reach the genital ridges, when a significant expansion of the PGC pool occurs (Tam and Snow, 1981; Anderson et al., 2000; Bowles and Koopman, 2010; Ewen and Koopman, 2010). To strengthen this notion, future experiments should investigate the role of TRIM71 in the proliferation of post-migratory PGCs in vivo. Additionally, the role of TRIM71 in the maintenance of the germline may extend beyond proliferation control, as previous studies have shown roles for TRIM71 not only in the control of proliferation (Chang et al., 2012; Chen et al., 2012, 2013; Ren et al., 2018; Hu et al., 2019; Torres-Fernández et al., 2019), but also during differentiation (Loedige et al., 2013; Worringer et al., 2014; Mitschka et al., 2015) and apoptosis (Nguyen et al., 2017; Chen et al., 2019; Hu et al., 2019) in developmental and oncogenic contexts. Importantly, a recent study described an impaired proliferation and an increased apoptosis in adult mice primary SSCs upon Trim71 KD (Du et al., 2020). Likewise, TRIM71 might also participate in the control of the massive apoptotic waves that mitotically arrested male PGCs/gonocytes undergo within the developing and neonatal gonads (Wang et al., 1998).
In summary, we have shown that germline-specific Trim71 cKO mice are infertile, with males predominantly displaying an SCO-like phenotype. We have identified TRIM71 variants in infertile men with severely impaired spermatogenesis, including a LoF variant in an SCO patient which was confirmed to abrogate TRIM71 expression and function. Trim71-associated infertility seems to originate during embryonic development, since the SCO-like phenotype was already apparent in neonatal P0.5 Trim71 cKO male mice and Trim71 expression in females is restricted to fetal gonads. Our in vitro assays consistently showed TRIM71-dependent changes in cell proliferation as well as in the expression of important PGC regulatory factors, suggesting that infertility in Trim71 cKO mice may be caused by an impaired proliferation of germ cells. Altogether, our work supports a novel role for TRIM71 in the embryonic development of the germline as well as of germline-derived tumors or GCT. Future unraveling of the molecular mechanisms by which TRIM71 governs germ cell biology will shed further light on the underlying causes of infertility. A deeper understanding of these processes will contribute to the development of new diagnostic and therapeutic strategies for reproductive medicine and for the treatment of GCT.
Methods
Mouse Generation
All animal experiments were conducted in a licensed animal facility in accordance with the German law on the protection of experimental animals (the German animal welfare act), and were approved (approval number 87-51.04.2011.A063) by local authorities of the state of Nordrhein-Westfalen (Landesamt für Natur-, Umwelt- und Verbraucherschutz NRW).
The generation of the full Trim71 KO mouse (Trim71fl/fl; Rosa26-CreERT2) was previously described (Mitschka et al., 2015). In order to generate a germline-specific Trim71 knockout mouse, wild type females with floxed Trim71 alleles (WT, Trim71fl/fl) were bred with male mice expressing the Cre recombinase under the control of the endogenous Nanos3 promoter in heterozygosity (Nanos3Cre/+) (Suzuki et al., 2008). The heterozygous Trim71–/+; Nanos3Cre/+ male offspring was then crossed with wild type Trim71+/+ females to produce Trim71 wild type animals with the Nanos3-Cre allele in heterozygosity (WT, Trim71+/+; Nanos3Cre/+), or with wild type Trim71fl/fl females to produce germline-specific heterozygous (cHET, Trim71–/+; Nanos3Cre/+) and germline-specific knockout (cKO, Trim71–/fl; Nanos3Cre/+) animals.
Genomic DNA Extraction and Genotyping
Genomic DNA was extracted from adult mice tail or ear biopsies or from mouse embryo yolk sacs by boiling the tissue in 50 mM NaOH for 20 min followed by pH neutralization of the lysate with 1/4 of 1 M Tris-Cl, pH 8.0. A three-primer strategy was used for PCR amplification of the Trim71 locus or the Nanos3 locus and fragments were resolved on a 2% agarose gel. Primers used are listed in Supplementary Table 3.
Epidydmal Sperm Counts
Cauda epididymis were isolated from 3-month-old male mice and transferred into 0.5–1 ml of PBS pre-warmed at 35–37°C. Sperm release was achieved by performing multiple cauda incisions followed by 10-min incubation at 35–37°C to allow the sperm swim out. The sperm suspension was then diluted in water (1:15-1:40) and sperm count was determined using a Neubauer hemocytometer.
Isolation of Spermatogonial Stem Cells From Mouse Testes
Testes were isolated from 2-month-old male mice, and seminiferous tubules were exposed by removing the tunica albuginea. The tubules of several testes were pooled and incubated in approximately 10 volumes of HBSS with calcium and magnesium containing 1 mg/ml collagenase Type IV and 200 to 500 μg/ml DNAse I, followed by incubation at 37°C under gentle agitation for 15 min. The tubules were then washed three times in 10 volumes of HBSS, followed by incubation at 37°C for 5 min in HBSS containing 0.25% trypsin and 150 μg/ml DNAse I under gentle agitation. Trypsinization was stopped by adding 20% FBS, and the cell suspension was filtered through a 40 μm pore size nylon filter. The filtrate was centrifuged at 1000 rpm for 5 min at 4°C and the cell pellet was resuspended in PBS before cell counting. Magnetic-activated cell sorting (MACS) was used to enrich THY1.2 + cells. To this end, 106 cells were stained with 0.2 μg FITC-labeled anti-THY1.2/CD90.2 antibody (BioLegend) for 15 min at 4°C. The cells were then washed in MACS-buffer (PBS supplemented with 2 mM EDTA and 0.5% FBS) and incubated in a 1:5 dilution of anti-FITC microbeads (Miltenyi) for 15 min at 4°C before a final washing in MACS buffer followed by centrifugation at 1000 rpm for 5 min at 4°C. The pellet was resuspended in 500 μl MACS buffer and loaded on an AutoMACS device (Miltenyi) with the program set for positive selection. The efficiency of enrichment was 8-fold as later controlled by flow cytometry. The different cell populations were then used for RNA extraction and qRT-PCR quantification.
H&E Staining of Mouse Testes Cross-Sections
PFA-fixed paraffin-embedded murine testes cross-sections were deparaffinized by incubation at 65°C for 15-20 min until the paraffin wax had melted, and washed in xylol twice for 10 min. Next, the sections were rehydrated by a descending ethanol dilution series (100% (2x), 95%, 90%, 80%, 70%) for 30 sec each and were kept in distilled water until staining. Cross-sections were stained in haematoxylin for 3 min and washed in cold running tap water for 3-5 min. Afterward, the sections were counterstained with 0.5% eosin for 3 min, removing excess dye by rinsing in cold running tap water for 1 min. Cross-sections were then dehydrated in an ascending ethanol dilution series [70%, 80%, 90%, 95%, 100% (2x)] and cleared in xylol twice for 2 min. Paraffin sections were then mounted with the xylene-based DPX mounting media for histology (Sigma-Aldrich). H&E stained sections were stored under the fume hood for 24 h before imaging by bright-field microscopy using the Zeiss Axio Lab.A1 microscope (Carl Zeiss).
Immunofluorescence Staining of Testes Cryosections
PFA-fixed murine testis cryosections stored at −20°C were defrosted and dried at room temperature for 15 min before their rehydration by washing in PBS and rinsing in distilled water. After drying the slides at RT for 10–15 min, cryosections were blocked (PBS supplemented with 1% BSA, 2% donkey serum and 0.3% Triton X-100) for 1 h at RT while covered with parafilm to prevent evaporation. Primary antibodies diluted in PBST (0.3% Triton X-100/PBS) supplemented with 1% BSA were added and incubated at 4°C overnight. On the next day, sections were washed three times for 5 min with PBST, and fluorescently conjugated secondary antibodies diluted in PBST supplemented with 1% BSA were added and incubated in the dark for 1 h at RT. The sections were then washed three times for 5 min with PBST and slides were mounted with Fluoromount-G containing DAPI and photobleaching inhibitors (SouthernBiotech). All steps were performed in a dark humid chamber to prevent evaporation and photobleaching. Stained sections were stored at 4°C for 24 h prior to imaging by immunofluorescence microscopy. Images were taken using the Zeiss Observer.Z1 epifluorescence microscope (Carl Zeiss) and the ZEN 2012 (blue edition) software (Carl Zeiss). Antibodies used are listed in Supplementary Table 4.
RNA Extraction and qRT-PCR Quantification
RNA was extracted from cell pellets using the Trizol-containing reagent peqGold TriFAST according to the manufacturer’s instructions (PeqLab). RNA pellets were resuspended in RNase-free water, and DNA digestion was performed prior to RNA quantification. 0.5–1 μg of RNA was reverse transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instructions. The cDNA was then diluted 1:5, and a relative quantification of specific genes was performed in a Bio-Rad qCycler using either TaqMan probes in iTaq Universal Probes Supermix or specific primer pairs in iTaq Universal SYBR Green Supermix (BioRad). Probes and Primers used are listed in Supplementary Table 3.
Protein Extraction and Western Blotting
Cell pellets were lyzed in RIPA buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1 mM Na3VO4, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM glycerophosphate) supplemented with protease inhibitors and protein lysates were pre-cleared by centrifugation and quantified using the BCA assay kit (Pierce) according to the manufacturer’s instructions. Protein lysates were then denatured by incubation with SDS buffer (12% glycerol, 60 mM Na2EDTA pH 8, 0.6% SDS, 0.003% bromophenol blue) for 10 min at 95°C and separated in SDS-PAGE gels in Laemmli buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). Proteins were then wet transferred to a nitrocellulose membrane in transfer buffer (25 mM Tris–HCl pH 7.6, 192 mM glycine, 20% methanol, 0.03% SDS) and membranes were blocked with 5% milk powder (w/v) diluted in 1 × TBST (50 mM Tris–HCl pH 7.6, 150 mM NaCl, 0.05% Tween-20) prior to overnight incubation at 4°C with the required primary antibodies. After washing the membrane three times with 1 × TBST, they were incubated with a suitable HRP-coupled secondary antibody for 1 h at RT, followed by three washing steps with 1 × TBST. Membranes were developed with the ECL substrate kit (Pierce) according to the manufacturer’s instructions. Antibodies used are listed in Supplementary Table 4.
Cell Culture and Transfections
Derivation of wild type murine ESCs (WT, Trim71fl/fl) from conditional Trim71 full KO mice (Trim71fl/fl; Rosa26-CreERT2) was previously described (Mitschka et al., 2015). Trim71 knockout murine ESCs (KO, Trim71–/–) were generated from wild type ESCs by addition of 500 nM of 4-hydroxytamoxifen in their culture media for 48 h, followed by further culture for 72 h to achieve full protein depletion. ESCs were cultured in 0.1% gelatin-coated dishes and maintained in 2i + LIF DMEM media (DMEM knockout media supplemented with 15% FCS, 1% penicillin-streptomycin, 0.1 mM NEAA, 2 mM L-GlutaMAX, 100 μM β-mercaptoethanol, 0.2% in-house produced LIF, 1 μM of MEK/ERK inhibitor PD0325091 and 3 μM of GSK-3 inhibitor CHIR99021).
The human hepatocellular carcinoma cell line HepG2 and the human embryonic carcinoma cell lines JKT-1, NCCIT, NTERA-2, TCam-2 and 2102EP were acquired from ATCC. HepG2, NCCIT and TCam-2 were cultured in RPMI 1640 media supplemented with 10% FBS and 1% penicillin–streptomycin antibiotic solution. JKT-1, NTERA -2 and 2012EP cells were cultured in DMEM media supplemented with 10% FBS and 1% penicillin-streptomycin antibiotic solution. DNA transfection in HEK293T cells was conducted with Lipofectamine2000 reagent following the manufacturer’s instructions and using 1 μg DNA per μl of Lipofectamine. RNA transfection in TCam-2 cells was performed with Lipofectamine RNAiMAX reagent following the manufacturer’s instructions and using 10 pmol siRNA per 2 μL of Lipofectamine. For proliferation assays in TCam-2 cells, 103 cells per well were seeded, transfected with the siRNAs in suspension (d0), and cell numbers were monitored over the course of several days (d1-d6).
In vitro Differentiation of ESCs Into PGCLCs
Embryonic stem cells growing under naïve pluripotency (d0) conditions on 0.1% gelatine-coated dishes and 2i + LIF N2B27 media were primed to EpiLCs (resembling post-implantation EpiSCs) for two days by plating 105 cells per well on 12-well plates coated with 20 μg/ml fibronectin in priming media (N2B27 media supplemented with 1% penicillin-streptomycin, 2 mM L-GlutaMAX, 1% KSR, 20 ng/ml Activin A and 12 ng/ml bFGF). After 24 h (d1), fresh priming media was provided. After 48 h (d2), EpiLCs were detached with Accutase Stem Cell Pro and 5000 cells per well were plated on a suspension U-bottom 96-well dish in specification media (GMEM media supplemented with 15% KSR, 1% penicillin-streptomycin, 0.1 mM NEAA, 2 mM L-GlutaMAX, 100 μM β-mercaptoethanol, 1 mM sodium pyruvate, 0.2% in-house produced LIF, 50 ng/ml EGF, 100 ng/ml SCF and 500 ng/ml BMP4). After 6 days in this media (d8), cells growing as spheroids were recovered, detached by trypsinization for 10 min at 37°C and either used for RNA extraction or double-stained with anti-ITGB3/CD61 and anti-SSEA-1/CD15 antibodies to determine the number of PGCLCs by flow cytometry. For staining, trypsinized cells were washed in 0.1% BSA/PBS, incubated with the antibodies diluted in 0.1% BSA/PBS for 15 min at 4°C in the dark and washed again in 0.1% BSA/PBS. Isotype control antibodies were used to control single stainings (data not shown). These protocols were adapted from a previous work (Hayashi et al., 2011). Antibodies used are listed in Supplementary Table 4.
Analysis of in vivo PGC Migration
E8.5-8.75 embryos were obtained from pregnant females with the first day of vaginal plug identification defined as 0.5 dpc. Embryos were dissected in PBS and the upper body was cut off and used for genotyping. The lower body was dissected to expose the hindgut and fixed in 4% PFA for 3 h at 4°C, followed by several washings in PBS. Embryos were then placed in 70% ethanol at 4°C for 1 h, washed once in PBS and stained with Fast Red TR and α-naphthyl phosphate (Sigma) following the manufacturer’s instructions to detect alkaline phosphatase-positive cells.
Generation of NCCIT Cells With TRIM71 Frameshift Mutations
Gene editing using the CRISPR/Cas9 system was carried out using the plasmid pSpCas9(BB)-2A-GFP (PX458), which was a kind gift from Feng Zhang (Addgene plasmid #48138), after the insertion of specific sgRNAs targeting TRIM71 RING (sgRNA ΔRING 5′-CACCGCTCGCAGACGCTCACGCTGT-3′) or NHL (sgRNA ΔNHL6 5′-CACCGCACAACGATCATTCCGCTGG-3′) domains. NCCIT cells were transfected with PX458 empty vector (EV), TRIM71 ΔRING vector or TRIM71 ΔNHL6 vector using Lipofectamine Stem Transfection Reagent (Invitrogen) following the manufacturer’s instructions. 48 h post- transfection, transfected (GFP-positive) cells were sorted by FACS and plated as a bulk population. Growth competition assays (see below) were performed with sorted bulk populations. Generation of single clones from the bulk populations was also performed for subsequent Sanger sequencing, western blot analysis, RNA extraction and qPCR measurements, and eFluo670 proliferation assays (see below).
Growth Competition Assays and NGS Allele Frequency Analysis via Illumina MiSeq
In order to analyze the growth behavior of CRISPR/Cas9-edited NCCIT cells, GFP-positive sorted bulk EV-transfected cells (WT) were mixed 1:1 with either TRIM71 sgRNA ΔRING-transfected cells or TRIM71 sgRNA ΔNHL6-transfected cells (d0) and cultured (2 × 105 cells per well in a 6-well plate) for three weeks to investigate the population dynamics. Samples (2 × 105 cells) were taken at d0, d3, d7, d14 and d21 for genomic DNA extraction. A double PCR strategy was then applied for the preparation of barcoded amplicons. In a first PCR reaction, the region around the CRISPR/Cas9-targeted site was amplified using site-specific forward and reverse primers with a common 5′ overhang. After PCR product cleaning, a second PCR was conducted with Illumina index primers which bind to the common 5′overhangs of the first primers and also contain unique barcode/index sequences followed by specific 5′ (P5) or 3′ (P7) adaptor sequences. PCR products were then gel-purified and sequenced on an Illumina MiSeq sequencing platform via amplification with common primers binding to P5/P7 adaptor sequences. Data analysis was performed using CRISPResso2 (Pinello et al., 2016) with the quality cut-off set at 30 and the minimum identity score for the alignment being adjusted to 50. After analysis of the sequencing data, single reads were categorized as wild type, frameshift mutations and in-frame mutations, and allele frequencies were calculated and displayed as percentages in a pie chart. Comparing changes in the distribution of single reads for wild type (EV) and frameshift mutants TRIM71 ΔRING or TRIM71 ΔNHL6 over time gives insights into the dynamics of each cell populations within the initially 1:1 mixed culture. Allele frequencies of pure wild type (EV) were also analyzed at d0 and d21 as a control. Analysis of allele frequencies of pure WT, TRIM71 ΔRING and TRIM71 ΔNHL6 populations before (d0 pure) and after mixing them 1:1 (d0 1:1) ensured that no PCR product was biased over another based on theoretically expected versus observed allele frequencies.
eFluor670 Proliferation Assays in NCCIT Cells
Cells were stained with the proliferation dye eFluor670 (eBiosciences) diluted to 5 μM in PBS (1 ml of PBS per 106 cells) for 10 min at 37°C in the dark. The labeling was stopped by addition of 4-5 volumes of cold FCS and incubation for 5 min on ice. Cells were washed once with complete growth medium and once with PBS before seeding them at different densities per condition in multi-well plates. A fraction of the cells was used to measure the initial fluorescence intensity (day 0) via flow cytometry. The loss of fluorescence intensity overtime was monitored by flow cytometry for 4 days after staining. Flow cytometry data was then analyzed with FlowJo. The number of cell divisions at the end of the experiment was calculated assuming a decrease of the median fluorescence intensity (MFI) by half upon each cell division with the following formula: Division Number = log2 [(MFIday0-MFIunstained)/(MFIday4-MFIunstained)]. The average cell cycle duration was estimated at the end of the experiment from the number of cell divisions.
Exome Sequencing Study Population
The study population consisted of 1025 individuals (MERGE cohort), of whom 908 otherwise healthy men presented with quantitative spermatogenic impairment. Patients attended the Centre of Reproductive Medicine and Andrology (CeRA) at the University Hospital Münster or the Clinic for Urology, Pediatric Urology and Andrology in Gießen. Neither individuals with previous chemo-/radiotherapy history nor patients with Klinefelter syndrome or AZF deletions, which represent known genetic causes of male infertility, were included in this study. A subgroup of 247 selected patients was used to investigate novel genetic causes of the SCO phenotype (SCO subcohort). They had previously undergone testicular biopsy and presented with complete and bilateral absence of germ cells. Specifically, two biopsies were taken from each testis, and tissue sections were done from each biopsy followed by the evaluation of all tubules in one random section per biopsy for exact determination of the phenotype. In addition to this subgroup, 89 male subjects with complete spermatogenesis also pertaining to the MERGE cohort were included in the study as controls. All individuals gave written informed consent and the study protocol has been approved by the appropriate ethics committees according to the Declaration of Helsinki (Münster: Kennzeichen 2010-578-f-S, Gießen: No. 26/11). As further controls, we utilized a Dutch cohort of 5784 proven fathers sequenced at the Radboudumc genome diagnostics center in Nijmegen, the Netherlands. These were healthy fathers of a child with severe developmental delay and all parents underwent routine exome sequencing for trio analysis. The fertility of the fathers is expected to be similar to an unselected sample of the population.
Exome Sequencing Data Generation and Analysis
In order to perform exome sequencing, genomic DNA was isolated from the subjects’ peripheral blood applying standard procedures. Target enrichment, library capture, exome sequencing and variant calling were performed as previously described (Wyrwoll et al., 2020). Briefly, DNA was enriched according to Agilent’s SureSelectQXT Target Enrichment for Illumina Multiplexed Sequencing Featuring Transposase-Based Library Prep Technology or Twist Bioscience’s Twist Human Core Exome protocol. For library capture, SureSelectXT Human All Exon Kits (Agilent) or Human Core Exome (Twist Bioscience) were used. Sequencing was then conducted on Illumina HiScanSQ®, Illumina NextSeq®500/550 or Illumina HiSeqX® systems.
Exome data analysis was conducted on 247 SCO patients using our platform Sciobase. We filtered genes based on the predicted effect of the variant on translated protein (including only stop-gain, frameshift, and splice donor/acceptor variants) and the allele frequency in the general population (including only rare variants (MAF ≤ 0.01) as listed in gnomAD genomic database (v2.1.11). This first filtering step resulted in the selection of 3997 genes. We then developed an application termed Haystack to facilitate the search for previously unknown causes of genetic diseases, technically applicable to exome data from patients with any clinical phenotype. In brief, Haystack aggregates information from multiple databases in an R Shiny application and allows for filtering based on the aggregated data, thus minimizing the manual effort required for candidate gene selection. All source code is publicly available at GitHub2. We used Haystack to filter variants from our SCO patients for quality (read depth ≥ 10, variant depth ≥ 4 and frequency ≥ 40%). The remaining 2870 genes were filtered again based on genotype and allele frequency in gnomAD (MAF ≤ 0.01 for homozygous variants and MAF ≤ 0.001 for heterozygous and X-linked variants) as well as on expression levels in the testes [including only genes whose expression in testis was above 50% of its maximum expression across all tissues according to the GTEx portal (v73)]. Last, genes with LoF variants present in individuals with intact spermatogenesis were excluded from the selected 949 genes, leaving a total of 721 genes with LoF variants specifically found in SCO patients. TRIM71 was found among these genes in a patient with an SCO phenotype (subject M364). This individual’s DNA was subjected to Sanger sequencing according to standard procedures in order to confirm his TRIM71 frameshift variant. PCR was performed using the primers: F: 5′-TGCAGGCCTAATCGATGCAT-3′; R: 5′-GAAGAAGCTGCGTTGCCCTC-3′.
For the statistical analysis of exome data, the number of alleles affected and unaffected by LoF variants in TRIM71 in all gnomAD subjects was compared to affected and unaffected alleles in the 908 patients with quantitative spermatogenic impairment (MERGE cohort) and the 247 SCO patients (SCO subcohort). A two-sided Fisher’s exact test was performed using the Woolf logit method.
In a subsequent step, exome data of the entire MERGE cohort and the Dutch father cohort was screened for rare (see MAF cutoffs above) missense variants in TRIM71 (NM_001039111). The Z score for TRIM71 was obtained from gnomAD database. Predicted pathogenicity was assessed using the scoring algorithms CADD (v14), PolyPhen (v25), SIFT (v26), and Mutation Taster (v67). All variants identified by our study have been submitted to ClinVar database (accession ID SUB9045938). Known genetic causes of azoospermia, such as chromosomal aberrations and AZF deletions were excluded in all infertile patients with LoF or missense variants in TRIM71. Furthermore, all patients carrying LoF or missense variants in TRIM71 (listed in Table 1) were evaluated for relevant variants in other genes (n = 181, Supplementary Table 2) that could explain the patients’ infertility. The majority of examined genes (n = 170, Supplementary Table 2A) were taken from a review on the genetics of male infertility (Oud et al., 2019). The included genes were selected based on associated phenotypes and had been rated with limited to definitive clinical evidence. The list was amended by adding 11 recently published genes (Supplementary Table 2B). Specifically, the patients’ exome data was screened for stop-gain, frameshift, and splice acceptor/donor variants as well as missense variants with a CADD score ≥ 20. Only rare variants (gnomAD MAF ≤ 0.01 for homozygous and MAF ≤ 0.001 for heterozygous variants) were taken into account. For recessive genes, variants were only considered if found in homozygous or likely compound-heterozygous state (two variants in the same gene in the same patient).
Luciferase Reporter Assays
HEK293T cells were co-transfected with the suitable psiCHECK2 dual luciferase plasmid (Promega) and the specified Flag-tagged TRIM71 variant construct in a 1:4 ratio. The psiCHECK2 plasmids contained a specific 3′UTR sequence downstream of the Renilla Luciferase ORF (either the CDKN1A 3′UTR Torres-Fernández et al., 2019 or an artificial 3′UTR with 8x Let-7 binding sites Torres-Fernández et al., 2021), while the Firefly Luciferase encoded in the same plasmid was used as a normalization control. Transfected cells were harvested 48 h post-transfection and Renilla and Firefly signals were measured consecutively with the use of a luminometer (MicroLumat Plus LB96V, Berthold) and the Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s instructions.
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: The exome sequencing data is part of the Reproductive Genomics (MERGE) study conducted by the Institute of Reproductive Genetics, University of Münster, and contains data mostly from patients attending the Centre of Reproductive Medicine and Andrology (CeRA), University Hospital Münster. Requests to access these datasets should be directed to FT, frank.tuettelmann@ukmuenster.de.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the State Medical Board and the Medical Faculty of the University of Münster. The subjects provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article. The animal study was reviewed and approved by Landesamt f r Natur-, Umwelt- und Verbraucherschutz NRW. Patients attended the Centre of Reproductive Medicine and Andrology (CeRA) at the University Hospital M nster or the Clinic for Urology, Pediatric Urology and Andrology in Gießen. All individuals gave written informed consent and the study protocol has been approved by the appropriate ethics committees according to the Declaration of Helsinki (Münster: Kennzeichen 2010-578-f-S, Gießen: No. 26/11).
Author Contributions
LT-F, JE, YP, and SM designed and performed experiments. LT-F and JE wrote the manuscript. SM and SS developed the germline-specific Trim71 KO mouse. YP, SM, SS, and LT-F performed in vivo and in vitro experiments. MH and YP performed NGS Illumina MiSeq. NN and SD provided testis single-cell RNAseq data. SK and DF provided samples and histological evaluation for patients from Münster and Gießen, respectively. JE and MW developed the variant filtering tool Haystack. JE performed bioinformatics analyses. MO provided genetic data for the cohort of Dutch fathers. WK, FT, HS, and CF supervised experimental design, data analysis and manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
LT-F held a stipend from Bayer AG. WK is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy EXC2151 – 390873048. This work was carried out within the frame of the German Research Foundation (DFG) funded Clinical Research Unit ‘Male Germ Cells: from Genes to Function’ (CRU326, grants to NN, CF, and FT).
Conflict of Interest
LT-F held a stipend which was donated by Bayer AG.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the Kolanus lab and the LIMES institute for general advice and discussion. We are indebted to all subjects having donated their DNA for research use in our studies. The technical work of Laura Struffert, Christina Burhöi and Judith Ryll is gratefully acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.658966/full#supplementary-material
Footnotes
- ^ https://gnomad.broadinstitute.org
- ^ https://github.com/MarWoes/haystack
- ^ https://gtexportal.org
- ^ https://cadd.gs.washington.edu/
- ^ http://genetics.bwh.harvard.edu/pph2
- ^ https://sift.bii.a-star.edu.sg
- ^ http://www.mutationtaster.org
References
Agarwal, A., Mulgund, A., Hamada, A., and Chyatte, M. R. (2015). A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 13:37.
Anderson, R., Copeland, T. K., Schöler, H., Heasman, J., and Wylie, C. (2000). The onset of germ cell migration in the mouse embryo. Mech. Dev. 91, 61–68. doi: 10.1016/s0925-4773(99)00271-3
Bowles, J., and Koopman, P. (2010). Sex determination in mammalian germ cells: extrinsic versus intrinsic factors. Reproduction 139, 943–958. doi: 10.1530/rep-10-0075
Chang, H.-M., Martinez, N. J., Thornton, J. E., Hagan, J. P., Nguyen, K. D., and Gregory, R. I. (2012). Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 3:923.
Chen, J., Lai, F., and Niswander, L. (2012). The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 26, 803–815. doi: 10.1101/gad.187641.112
Chen, Y., Hao, Q., Wang, J., Li, J., Huang, C., Zhang, Y., et al. (2019). Ubiquitin ligase TRIM71 suppresses ovarian tumorigenesis by degrading mutant p53. Cell Death Dis. 10, 1–14.
Chen, Y. L., Yuan, R. H., Yang, W. C., Hsu, H. C., and Jeng, Y. M. (2013). The stem cell E3-ligase Lin-41 promotes liver cancer progression through inhibition of microRNA-mediated gene silencing. J. Pathol. 229, 486–496. doi: 10.1002/path.4130
Cuevas, E., Rybak-Wolf, A., Rohde, A. M., Nguyen, D. T. T., and Wulczyn, F. G. (2015). Lin41/Trim71 is essential for mouse development and specifically expressed in postnatal ependymal cells of the brain. Front. Cell Dev. Biol. 3:20. doi: 10.3389/fcell.2015.00020
Du, G., Wang, X., Luo, M., Xu, W., Zhou, T., Wang, M., et al. (2020). mRBPome capture identifies the RNA binding protein TRIM71, an essential regulator of spermatogonial differentiation. Development 147:dev184655. doi: 10.1242/dev.184655
Ecsedi, M., and Grosshans, H. (2013). LIN-41/TRIM71: emancipation of a miRNA target. Genes Dev. 27, 581–589. doi: 10.1101/gad.207266.112
Elsea, S. H., and Lucas, R. E. (2002). The mousetrap: what we can learn when the mouse model does not mimic the human disease. ILAR J. 43, 66–79. doi: 10.1093/ilar.43.2.66
Enders, G. C., and May, J. J. (1994). Developmentally regulated expression of a mouse germ cell nuclear antigen examined from embryonic day 11 to adult in male and female mice. Dev. Biol. 163, 331–340. doi: 10.1006/dbio.1994.1152
Ewen, K. A., and Koopman, P. (2010). Mouse germ cell development: from specification to sex determination. Mol. Cell. Endocrinol. 323, 76–93. doi: 10.1016/j.mce.2009.12.013
Foster, D. J., Chang, H. M., Haswell, J. R., Gregory, R. I., and Slack, F. J. (2020). TRIM71 binds to IMP1 and is capable of positive and negative regulation of target RNAs. Cell Cycle 19, 2314–2326. doi: 10.1080/15384101.2020.1804232
Fujiwara, Y., Komiya, T., Kawabata, H., Satot, M., Fujimotot, H., Furusawa, M., et al. (1994). Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Dev. Biol. 91, 12258–12262. doi: 10.1073/pnas.91.25.12258
Furey, C. G., Choi, J., Jin, S. C., Zeng, X., Timberlake, A. T., Nelson-Williams, C., et al. (2018). De novo mutation in genes regulating neural stem cell fate in human congenital hydrocephalus. Neuron 99, 302.e4–314.e4.
Gallardo, T., Shirley, L., John, G. B., and Castrillon, D. H. (2007). Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413–417. doi: 10.1002/dvg.20310
Gassei, K., and Orwig, K. E. (2013). SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS One 8:e53976. doi: 10.1371/journal.pone.0053976
Gaytan, F., Sangiao-Alvarellos, S., Manfredi-Lozano, M., García-Galiano, D., and Ruiz-Pino, F. (2013). Distinct expression patterns predict differential roles of the mirna-binding proteins, lin28 and lin28b, in the mouse testis: studies during postnatal development and in a model of hypogonadotropic hypogonadism. Endocrinology 154, 1321–1336. doi: 10.1210/en.2012-1745
Ginsburg, M., Snow, M. H. L., and Mclaren, A. (1990). Primordial germ cells in the mouse embryo during gastrulation. Development 110, 521–528.
Grabole, N., Tischler, J., Hackett, J. A., Kim, S., Tang, F., Leitch, H. G., et al. (2013). Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation. EMBO Rep. 14, 629–637. doi: 10.1038/embor.2013.67
Guo, G., Yang, J., Nichols, J., Hall, J. S., Eyres, I., Mansfield, W., et al. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069. doi: 10.1242/dev.030957
Han, X., Wang, R., Zhou, Y., Fei, L., Sun, H., Lai, S., et al. (2018). Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091.e17–1107.e17.
Hayashi, K., Ohta, H., Kurimoto, K., Aramaki, S., and Saitou, M. (2011). Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532. doi: 10.1016/j.cell.2011.06.052
Hermann, B. P., Cheng, K., Singh, A., Roa-De La Cruz, L., Mutoji, K. N., Chen, I. C., et al. (2018). The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 25, 1650.e8–1667.e8.
Hu, Q., Ye, Y., Chan, L. C., Li, Y., Liang, K., Lin, A., et al. (2019). Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat. Immunol. 20, 835–851.
Irie, N., Weinberger, L., Tang, W. W. C., Kobayashi, T., Viukov, S., Manor, Y. S., et al. (2015). SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253–268. doi: 10.1016/j.cell.2014.12.013
Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alföldi, J., Wang, Q., et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443.
Kehler, J., Tolkunova, E., Koschorz, B., Pesce, M., Gentile, L., Boiani, M., et al. (2004). Oct4 is required for primordial germ cell survival. EMBO Rep. 5, 1078–1083. doi: 10.1038/sj.embor.7400279
Koc, G., Ozdemir, A. A., Girgin, G., Akbal, C., Kirac, D., Avcilar, T., et al. (2019). Male infertility in Sertoli cell-only syndrome: an investigation of autosomal gene defects. Int. J. Urol. 26, 292–298. doi: 10.1111/iju.13863
Koshimizu, U., Nishioka, H., Watanabe, D., Dohmae, K., and Nishimune, Y. (1995). Characterization of a novel spermatogenic cell antigen specific for early stages of germ cells in mouse testis. Mol. Reprod. Dev. 40, 221–227. doi: 10.1002/mrd.1080400211
Krausz, C., Riera-Escamilla, A., Moreno-Mendoza, D., Holleman, K., Cioppi, F., Algaba, F., et al. (2020). Genetic dissection of spermatogenic arrest through exome analysis: clinical implications for the management of azoospermic men. Genet. Med. 22, 1956-1966.
Kumari, P., Aeschimann, F., Gaidatzis, D., Keusch, J. J., Ghosh, P., Neagu, A., et al. (2018). Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition. Nat. Commun. 9:1549.
Kurimoto, K., Yabuta, Y., Ohinata, Y., Shigeta, M., Yamanaka, K., and Saitou, M. (2008). Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 22, 1617–1635. doi: 10.1101/gad.1649908
Lawson, K. A., Dunn, N. R., Roelen, B. A. J., Zeinstra, L. M., Davis, A. M., Wright, C. V. E., et al. (1999). Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 13, 424–436. doi: 10.1101/gad.13.4.424
Lee, J. Y., Dada, R., Sabanegh, E., Carpi, A., and Agarwal, A. (2011). Role of genetics in azoospermia. Urology 77, 598–601. doi: 10.1016/j.urology.2010.10.001
Liao, H.-F., Kuo, J., Lin, H.-H., and Lin, S.-P. (2016). Isolation of THY1+ undifferentiated spermatogonia from mouse postnatal testes using magnetic-activated cell sorting (MACS). Bio Protocol 6:2072.
Loedige, I., Gaidatzis, D., Sack, R., Meister, G., and Filipowicz, W. (2013). The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic Acids Res. 41, 518–532. doi: 10.1093/nar/gks1032
Looijenga, L. H. J., Hersmus, R., De Leeuw, B. H. C. G. M., Stoop, H., Cools, M., Oosterhuis, J. W., et al. (2010). Gonadal tumours and DSD. Best Pract. Res. Clin. Endocrinol. Metab. 24, 291–310. doi: 10.1016/j.beem.2009.10.002
Mitschka, S., Ulas, T., Goller, T., Schneider, K., Egert, A., Mertens, J., et al. (2015). Co-existence of intact stemness and priming of neural differentiation programs in mES cells lacking Trim71. Sci. Rep. 5:11126.
Mitsunaga, S., Odajima, J., Yawata, S., Shioda, K., Owa, C., Isselbacher, K. J., et al. (2017). Relevance of iPSC-derived human PGC-like cells at the surface of embryoid bodies to prechemotaxis migrating PGCs. Proc. Natl. Acad. Sci. U.S.A. 114, E9913–E9922.
Müller, M. R., Skowron, M. A., Albers, P., and Nettersheim, D. (2020). Molecular and epigenetic pathogenesis of germ cell tumors. Asian J. Urol. 2, 153–167.
Mundlos, S., Pelletier, J., Darveau, A., Bachmann, M., Winterpacht, A., and Zabel, B. (1993). Nuclear localization of the protein encoded by the Wilms’ tumor gene WT1 in embryonic and adult tissues. Development 119, 1329–1341.
Nettersheim, D., Arndt, I., Sharma, R., Riesenberg, S., Jostes, S., Schneider, S., et al. (2016a). The cancer/testis-antigen PRAME supports the pluripotency network and represses somatic and germ cell differentiation programs in seminomas. Br. J. Cancer 115, 454–464. doi: 10.1038/bjc.2016.187
Nettersheim, D., Heimsoeth, A., Jostes, S., Schneider, S., Fellermeyer, M., Hofmann, A., et al. (2016b). SOX2 is essential for in vivo reprogramming of seminoma-like TCam-2 cells to an embryonal carcinoma-like fate. Oncotarget 7, 47095–47110. doi: 10.18632/oncotarget.9903
Nettersheim, D., Jostes, S., Schneider, S., and Schorle, H. (2016c). Elucidating human male germ cell development by studying germ cell cancer. Reproduction 152, R101–R113.
Nguyen, D. T. T., Richter, D., Michel, G., Mitschka, S., Kolanus, W., Cuevas, E., et al. (2017). The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differentiation pathways during stem cell differentiation. Cell Death. Differ. 24, 1063–1078. doi: 10.1038/cdd.2017.54
Ohinata, Y., Payer, B., O’Carroll, D., Ancelin, K., Ono, Y., Sano, M., et al. (2005). Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213. doi: 10.1038/nature03813
Oosterhuis, J. W., and Looijenga, L. H. J. (2005). Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer 5, 210–222. doi: 10.1038/nrc1568
Oud, M. S., Volozonoka, L., Smits, R. M., Vissers, L. E. L. M., Ramos, L., and Veltman, J. A. (2019). A systematic review and standardized clinical validity assessment of male infertility genes. Hum. Reprod. 34, 932–941. doi: 10.1093/humrep/dez022
Pierce, J. L., Frazier, A. L., and Amatruda, J. F. (2018). Pediatric germ cell tumors: a developmental perspective. Adv. Urol. 2018:9059382.
Pinello, L., Canver, M. C., Hoban, M. D., Orkin, S. H., Kohn, D. B., Bauer, D. E., et al. (2016). Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 34, 695–697. doi: 10.1038/nbt.3583
Raman, J. D., Nobert, C. F., and Goldstein, M. (2005). Increased incidence of testicular cancer in men presenting with infertility and abnormal semen analysis. J. Urol. 174, 1819–1822. doi: 10.1097/01.ju.0000177491.98461.aa
Ravi, R. K., Walton, K., and Khosroheidari, M. (2018). Miseq: A. (next)generation sequencing platform for genomic analysis. Methods Mol. Biol. 1706, 223–232. doi: 10.1007/978-1-4939-7471-9_12
Ren, H., Xu, Y., Wang, Q., Jiang, J., Hui, L., Zhang, Q., et al. (2018). E3 ubiquitin ligase tripartite motif-containing 71 promotes the proliferation of non-small cell lung cancer through the inhibitor of kappaB-α/nuclear factor kappaB pathway. Oncotarget 9, 10880–10890. doi: 10.18632/oncotarget.19075
Rosner, M. H., Vigano, M. A., Ozato, K., Timmons, P. M., Poirie, F., Rigby, P. W. J., et al. (1990). A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature 345, 686–692. doi: 10.1038/345686a0
Rybak, A., Fuchs, H., Hadian, K., Smirnova, L., Wulczyn, E. A., Michel, G., et al. (2009). The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol. 11, 1411–1420. doi: 10.1038/ncb1987
Schilit, S. L. P., Menon, S., Friedrich, C., Kammin, T., Wilch, E., Hanscom, C., et al. (2019). SYCP2 translocation-mediated dysregulation and frameshift variants cause human male infertility. Am. J. Hum. Genet. 106, 41–57. doi: 10.1016/j.ajhg.2019.11.013
Schulman, B. R. M., Esquela-Kerscher, A., and Slack, F. J. (2005). Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev. Dyn. 234, 1046–1054.
Schulman, B. R. M., Liang, X., Stahlhut, C., Delconte, C., Stefani, G., and Slack, F. J. (2008). The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle 7, 3935–3942.
Seisenberger, S., Andrews, S., Krueger, F., Arand, J., Walter, J., Santos, F., et al. (2012). The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862.
Seki, Y. (2018). PRDM14 is a unique epigenetic regulator stabilizing transcriptional networks for pluripotency. Front. Cell Dev. Biol. 6:12. doi: 10.3389/fcell.2018.00012
Slack, F. J., Basson, M., Liu, Z., Ambros, V., Horvitz, H. R., and Ruvkun, G. (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5, 659–669.
Soh, Y. Q. S., Junker, J. P., Gill, M. E., Mueller, J. L., van Oudenaarden, A., and Page, D. C. (2015). A gene regulatory program for meiotic prophase in the fetal ovary. PLoS Genet. 11:e1005531.
Spike, C. A., Coetzee, D., Eichten, C., Wang, X., Hansen, D., and Greenstein, D. (2014). The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of caenorhabditis elegans oocytes. Genetics 198, 1535–1558.
Sugawa, F., Araúzo-Bravo, M. J., Yoon, J., Kim, K. P., Aramaki, S., Wu, G., et al. (2015). Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 34, 1009–1024.
Suzuki, H., Tsuda, M., Kiso, M., and Saga, Y. (2008). Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev. Biol. 318, 133–142.
Tam, P. P. L., and Snow, M. H. L. (1981). Proliferation and migration of primordial germ cells during compensatory growth in mouse embryos. J. Embryol. Exp. Morphol. Vol. 64, 133–147.
Tanaka, H., Pereira, L. A. V. D., Nozaki, M., Tsuchida, J., Sawada, K., Mori, H., et al. (1998). A germ cell-specific nuclear antigen recognized by a monoclonal antibody raised against mouse testicular germ cells. Int. J. Androl. 20, 361–366.
Tanaka, S. S., Toyooka, Y., Akasu, R., Katoh-Fukui, Y., Nakahara, Y., Suzuki, R., et al. (2000). The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 14, 841–853.
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102.
Torres-Fernández, L. A., Jux, B., Bille, M., Port, Y., Schneider, K., Geyer, M., et al. (2019). The mRNA repressor TRIM71 cooperates with Nonsense-Mediated Decay factors to destabilize the mRNA of CDKN1A/p21. Nucleic Acids Res. 47:1057.
Torres-Fernández, L. A., Mitschka, S., Ulas, T., Dahm, K., Becker, M., Händler, K., et al. (2021). The stem cell-specific protein TRIM71 inhibits maturation and activity of the pro-differentiation miRNA let-7 via two independent molecular mechanisms. bioRxiv [Preprint]. doi: 10.1101/2021.01.29.428807v1
Tsuda, M., Sasaoka, Y., Kiso, M., Abe, K., Haraguchi, S., Kobayashi, S., et al. (2003). Conserved role of nanos proteins in germ cell development. Science 301, 1239–1241.
Tsukamoto, T., Gearhart, M. D., Spike, C. A., Huelgas-Morales, G., Mews, M., Boag, P. R., et al. (2017). LIN-41 and OMA ribonucleoprotein complexes mediate a translational repression-to-activation switch controlling oocyte meiotic maturation and the oocyte-to-embryo transition in Caenorhabditis elegans. Genetics 206, 2007–2039.
Tüttelmann, F., Ruckert, C., and Röpke, A. (2018). Disorders of spermatogenesis. Medizinische Genet. 30, 12–20.
Tüttelmann, F., Simoni, M., Kliesch, S., Ledig, S., Dworniczak, B., Wieacker, P., et al. (2011). Copy number variants in patients with severe oligozoospermia and sertoli-cell-only syndrome. PLoS One 6:e19426. doi: 10.1371/journal.pone.0019426
Van Der Bijl, N., Röpke, A., Biswas, U., Wöste, M., Jessberger, R., Kliesch, S., et al. (2019). Mutations in the stromal antigen 3 (STAG3) gene cause male infertility due to meiotic arrest. Hum. Reprod. 34, 2112–2119.
Veillard, A. C., Marks, H., Bernardo, A. S., Jouneau, L., Laloë, D., Boulanger, L., et al. (2014). Stable methylation at promoters distinguishes epiblast stem cells from embryonic stem cells and the in vivo epiblasts. Stem Cells Dev. 23, 2014–2029.
Wang, J.-Q., and Cao, W.-G. (2016). Key signaling events for committing mouse pluripotent stem cells to the germline fate1. Biol. Reprod. 94, 1–9.
Wang, R. A., Nakane, P. K., and Koji, T. (1998). Autonomous cell death of mouse male germ cells during fetal and postnatal period. Biol. Reprod. 58, 1250–1256.
Welte, T., Tuck, A. C., Papasaikas, P., Carl, S. H., Flemr, M., Knuckles, P., et al. (2019). The RNA hairpin binder TRIM71 modulates alternative splicing by repressing MBNL1. Genes Dev. 33, 1221–1235.
West, J. A., Viswanathan, S. R., Yabuuchi, A., Cunniff, K., Takeuchi, A., Park, I. H., et al. (2009). A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature 460, 909–913.
Wiel, L., Baakman, C., Gilissen, D., Veltman, J. A., Vriend, G., and Gilissen, C. (2019). MetaDome: pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 40, 1030–1038.
Worringer, K. A., Rand, T. A., Hayashi, Y., Sami, S., Takahashi, K., Tanabe, K., et al. (2014). The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14, 40–52.
Wyrwoll, M. J., Temel, S. G., Nagirnaja, L., Oud, M., Lopes, A., van der Heijden, G. W., et al. (2020). Bi-allelic mutations in M1AP are a frequent cause of meiotic arrest and severely impaired spermatogenesis leading to male infertility. Am. J. Hum. Genet. 107, 342–351.
Yamaji, M., Seki, Y., Kurimoto, K., Yabuta, Y., Yuasa, M., Shigeta, M., et al. (2008). Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 40, 1016–1022.
Yatsenko, A. N., Georgiadis, A. P., Ropke, A., Berman, A. J., Jaffe, T., Olszewska, M., et al. (2015). X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med. 372, 2097–2107.
Zhao, L., Wang, C., Lehman, M. L., He, M., An, J., Svingen, T., et al. (2018). Transcriptomic analysis of mRNA expression and alternative splicing during mouse sex determination. Mol. Cell. Endocrinol. 478, 84–96.
Zheng, K., Wu, X., Kaestner, K. H., and Wang, P. J. (2009). The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev. Biol. 9:38.
Keywords: TRIM71/LIN-41, infertility/sterility, azoospermia, Sertoli cell-only (SCO) phenotype, primordial germ cells (PGCs), mammalian gonad development, germ cell tumors (GCT), Haystack
Citation: Torres-Fernández LA, Emich J, Port Y, Mitschka S, Wöste M, Schneider S, Fietz D, Oud MS, Di Persio S, Neuhaus N, Kliesch S, Hölzel M, Schorle H, Friedrich C, Tüttelmann F and Kolanus W (2021) TRIM71 Deficiency Causes Germ Cell Loss During Mouse Embryogenesis and Is Associated With Human Male Infertility. Front. Cell Dev. Biol. 9:658966. doi: 10.3389/fcell.2021.658966
Received: 26 January 2021; Accepted: 30 March 2021;
Published: 13 May 2021.
Edited by:
Takuya Wakai, Okayama University, JapanReviewed by:
Hiroshi Ohta, Kyoto University, JapanHarry Leitch, London Institute of Medical Sciences, Medical Research Council, United Kingdom
Copyright © 2021 Torres-Fernández, Emich, Port, Mitschka, Wöste, Schneider, Fietz, Oud, Di Persio, Neuhaus, Kliesch, Hölzel, Schorle, Friedrich, Tüttelmann and Kolanus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Waldemar Kolanus, wkolanus@uni-bonn.de; Frank Tüttelmann, frank.tuettelmann@ukmuenster.de
†These authors have contributed equally to this work
 Lucia A. Torres-Fernández
Lucia A. Torres-Fernández Jana Emich
Jana Emich Yasmine Port1†
Yasmine Port1†  Marius Wöste
Marius Wöste Manon S. Oud
Manon S. Oud Sara Di Persio
Sara Di Persio Nina Neuhaus
Nina Neuhaus Sabine Kliesch
Sabine Kliesch Michael Hölzel
Michael Hölzel Hubert Schorle
Hubert Schorle Corinna Friedrich
Corinna Friedrich Frank Tüttelmann
Frank Tüttelmann Waldemar Kolanus
Waldemar Kolanus