Regulation of Skeletal Muscle Atrophy in Cachexia by MicroRNAs and Long Non-coding RNAs
- 1Guangdong Traditional Medical and Sports Injury Rehabilitation Research Institute, Guangdong Second Provincial General Hospital, Guangzhou, China
- 2Department of Radiology, The Third Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
Skeletal muscle atrophy is a common complication of cachexia, characterized by progressive bodyweight loss and decreased muscle strength, and it significantly increases the risks of morbidity and mortality in the population with atrophy. Numerous complications associated with decreased muscle function can activate catabolism, reduce anabolism, and impair muscle regeneration, leading to muscle wasting. microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), types of non-coding RNAs, are important for regulation of skeletal muscle development. Few studies have specifically identified the roles of miRNAs and lncRNAs in cellular or animal models of muscular atrophy during cachexia, and the pathogenesis of skeletal muscle wasting in cachexia is not entirely understood. To develop potential approaches to improve skeletal muscle mass, strength, and function, a more comprehensive understanding of the known key pathophysiological processes leading to muscular atrophy is needed. In this review, we summarize the known miRNAs, lncRNAs, and corresponding signaling pathways involved in regulating skeletal muscle atrophy in cachexia and other diseases. A comprehensive understanding of the functions and mechanisms of miRNAs and lncRNAs during skeletal muscle wasting in cachexia and other diseases will, therefore, promote therapeutic treatments for muscle atrophy.
Introduction
Cachexia is a complex metabolic condition accompanied by progressive bodyweight loss, skeletal muscle atrophy, and decreased muscle strength. Muscle atrophy is an important factor associated with physical disability, poor quality of life, and increased mortality (Prado et al., 2008; Fearon et al., 2011). Cachexia is common in patients with certain cancers and other diseases in the advanced stages, such as patients with intensive care unit (ICU)-acquired (Garros et al., 2017) weakness or chronic obstructive pulmonary disease (COPD) (Man et al., 2009), chronic kidney disease (CKD) (Zhang et al., 2019), and heart failure (Anker et al., 1997). Patients with cachexia are less tolerant or responsive to interventions, which limits the treatment options and their efficacy, for example, the dose-limiting toxicity of radiation and chemotherapy in patients with cancer cachexia (de Castro et al., 2019).
Skeletal muscle atrophy in cachexia is due to a disruption in the balance between protein synthesis and degradation, which can be partially, but not fully reversed by nutritional support (Baracos et al., 2018; de Castro et al., 2019). Reduced protein levels result from decreased protein synthesis, increased degradation, and abnormalities in apoptosis or autophagy (Lok, 2015). Protein degradation pathways mainly include ubiquitin-proteasome pathway (UPP) and autophagy-lysosomal pathway (ALP). The ubiquitin-proteasome pathway is an active protein degradation pathway that degrades ubiquitinated proteins in cells. This way is mainly composed of ubiquitin, ubiquitin activating enzyme, ubiquitin conjugating enzyme, ubiquitin ligase (ubiquitinprotein ligase, E3), protease and its substrate. E3 specifically regulates target protein degradation by identifying and binding specific target protein sequences. In skeletal muscle, there are two muscle-specific E3 ligases: muscular atrophy fbox-1 protein (MAFbx/Atrogin-1) and muscle-specific RING finger protein -1 (MuRF-1). Atrogin-1 and MuRF-1 are key proteins leading to muscle protein degradation (Rom and Reznick, 2016). Models of muscle atrophy demonstrate a consistent increase in the expression level of Atrogin-1 and MuRF-1. Autophagy is a highly conserved process in eukaryotes that occurs in the cytoplasm, where excess or abnormal organelles are transported to lysosomes for degradation. There are three main types of autophagy in the cell, namely molecular chaperone-mediated autophagy, microautophagy and macrophage. And in our previous study, CoCl2-mimicked hypoxia induces autophagy in skeletal C2C12 myotubes, and increases the expression of muscle-specific ubiquitin ligase Atrogin-1, which reveals an important link between autophagy and muscle atrophy (Chen et al., 2017).
Whereas proteins are thought to be the dominant regulators of cell functions, a variety of non-protein-coding processes also operate. It is increasingly clear that the non-protein-coding parts of the genome are important for development, physiology, and disease progression (Sonkoly et al., 2005; Faghihi et al., 2008). microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are vital non-coding RNAs (ncRNAs). Studies have shown that miRNAs and lncRNAs may be associated with cancer (Ramalho-Carvalho et al., 2017), as well as neurological (Faghihi et al., 2008), cardiovascular (Ponnusamy et al., 2019), and other diseases (Iannone et al., 2020).
miRNAs and lncRNAs are also reported to play critical roles in muscle development and other muscle-related diseases, such as skeletal muscle wasting in cachexia (Chen et al., 2006, 2010; Li et al., 2017). miR-1 (Chen et al., 2006), miR-133 (Chen et al., 2006), miR-206 (Chen et al., 2010), and miR-29b (Li et al., 2017) can modulate muscle development and wasting. Our previous studies showed that 2,922 lncRNAs are differentially regulated during C2C12 differentiation, and these lncRNAs may be involved in multiple mechanisms regulating gene expression (Chen et al., 2018). In recent reports, linc-MD1 was shown to play a vital role through competing endogenous RNA (ceRNA) (Cesana et al., 2011), whereas lincRNA Yam-1 acted in cis (Lu et al., 2013), and lncRNA MUNC functioned in trans during skeletal muscle development (Cichewicz et al., 2018). A systematic review of our groups provided a summary of the known functions and mechanisms of lncRNAs in skeletal muscle development (Chen et al., 2020). However, 4,409 lncRNAs are differentially regulated by CoCl2 (Chen et al., 2019), which triggers atrophy in skeletal C2C12 myotubes (Chen et al., 2017), indicating the potential involvement of lncRNAs in muscle wasting.
To develop approaches to improve skeletal muscle mass, strength, and function, a more comprehensive understanding of the key pathophysiological processes leading to muscular atrophy is needed. Here, we summarize the known miRNAs and lncRNAs regulating skeletal muscle wasting in cachexia and other diseases. A comprehensive understanding of the functions and mechanisms of miRNAs and lncRNAs during muscle atrophy in cachexia will help identify potential therapeutic approaches for muscle atrophy.
miRNAs Involved in Skeletal Muscle Atrophy
Although the mechanism of muscular atrophy has been investigated extensively, the key processes driving muscular atrophy in different diseases requires further clarification. miRNAs are short ncRNA molecules (18–25 nucleotides) that are vital negative regulators of gene expression. They fine-tune gene expression via post-transcriptional mechanisms, leading to inhibition of translation or degradation of target mRNAs, and are involved in many biological pathways. miRNAs play important roles in maintaining skeletal muscle homeostasis by regulating muscle metabolism, growth, and regeneration. Muscle-specific miRNAs have emerged as significant regulators of muscle wasting.
miRNAs That Promote Skeletal Muscle Atrophy
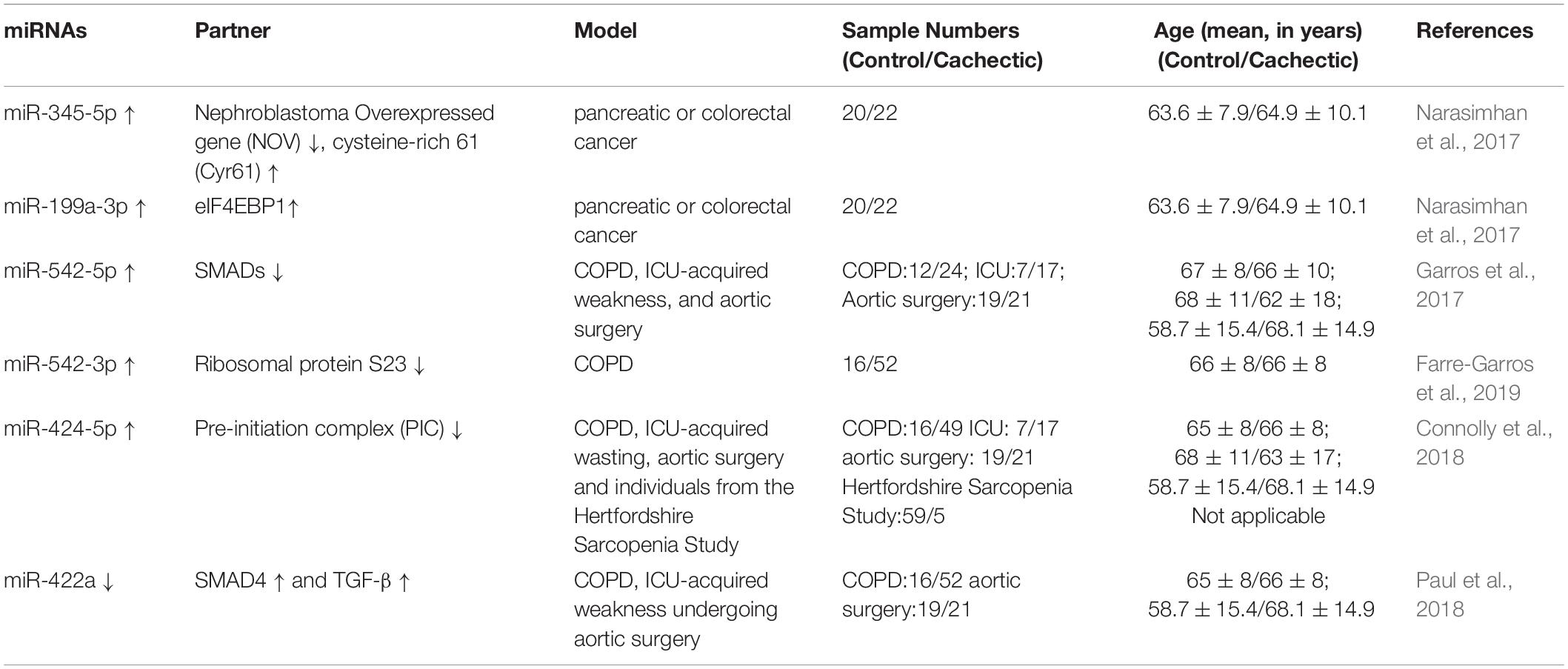
Emerging studies have demonstrated that certain miRNAs promote muscle atrophy via the PI3K/Akt/mTOR signaling pathway (Figure 1 and Tables 1, 2). miR-29b promotes atrophy by targeting insulin-like growth factor 1 (IGF-1) and PI3K in response to different atrophic stimuli in cells and mouse models. Inhibition of miR-29b attenuates atrophy induced by dexamethasone (Dex), tumor necrosis factor (TNF)-α, and H2O2 treatment (Li et al., 2017). It was reported that miR-345-5p downregulated nephroblastoma overexpressed (NOV) and upregulated cysteine-rich 61 (Cyr61) in human skeletal muscle with cancer cachexia. NOV and Cyr61 are involved in the IGF-1, Akt, and mTOR pathways which promote protein synthesis, thus miR-345-5p may lead to muscle atrophy by suppressing protein synthesis (Narasimhan et al., 2017). In addition, miR-18a blocks PI3K/Akt signaling by direct inhibition of IGF-1 expression in a 3’UTR-dependent manner in C2C12 myotubes and in mice, and it also decreases the phosphorylation of Akt and FoxO3, thus increases the expression of Atrogin-1, MuRF-1, and cathepsin L, and eventually leads to myotube atrophy (Liu et al., 2017).

Figure 1. Functional miRNAs involved in promoting skeletal muscle wasting in cachexia. miRNAs are represented in blue; molecules directly targeted by the miRNAs are shown in pink; and other signaling molecules involved in muscle wasting are shown in purple.
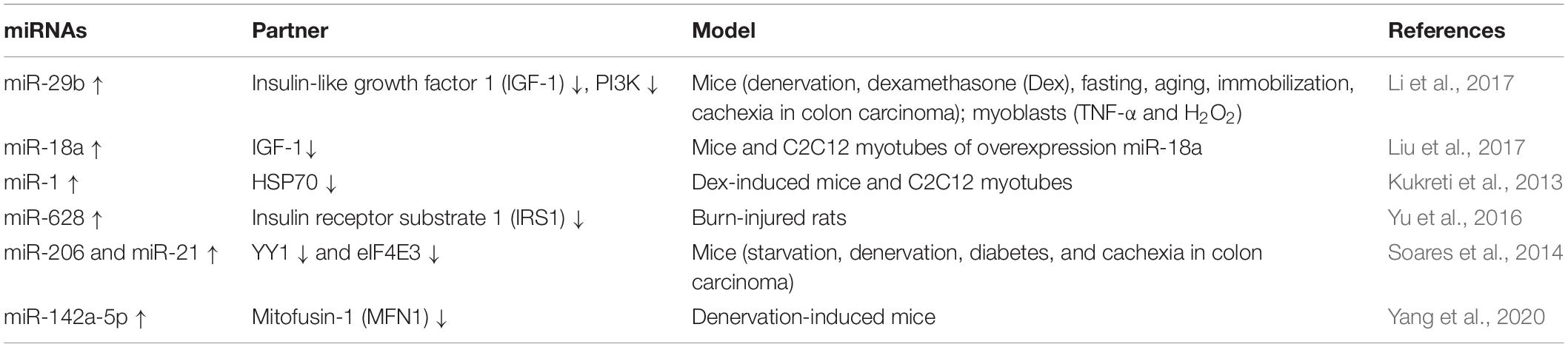
Table 1. Functional miRNAs involved in promoting skeletal muscle wasting in cell or rodent cachexia.
In addition to the PI3K/Akt/mTOR pathway, two miRNAs can enhance muscle wasting via Akt/FoxO signaling (Figure 1 and Table 1). Increasing miR-1 expression during Dex-induced atrophy reduces the level of HSP70, which can result in decreased phosphorylation of Akt, enhanced activation of FoxO3, upregulation of Atrogin-1 and MuRF-1, and progression of skeletal muscle atrophy (Kukreti et al., 2013). miR-628 promotes burn-induced skeletal muscle atrophy via targeting insulin receptor substrate 1 (IRS1). Rats subjected to burn injury exhibited skeletal muscle atrophy, significantly decreased IRS1 protein expression, and significantly increased skeletal muscle cell apoptosis. miR-628, a potential regulator of IRS1 protein translation, was also clearly elevated after the burn injury. In experiments in L6 myocytes, apoptosis was accelerated after induction of miR-628 expression, and the protein levels of IRS1 and p-Akt, involved in the IRS1/Akt/FoxO pathway, were decreased significantly. The levels of the apoptosis-related proteins FoxO3 and cleaved caspase 3 were also increased after induction of miR-628 expression (Yu et al., 2016).
Besides the above signaling pathways, some miRNAs facilitate cachexia muscle atrophy by regulating eukaryotic initiation factor 4E (eIF4E), one of the most critical factors regulating ribosome assembly and protein synthesis (Figure 1 and Tables 1, 2). miR-206 enhances wasting by decreasing the action of eIF4E3, and miR-21 inhibits the expression of transcription factor YY1 and eIF4E3, thus promoting muscle wasting in four different models: starvation, denervation, diabetes, and cancer cachexia induced by colon carcinoma (Soares et al., 2014). YY1 is reported to play an important role in the maintenance of mitochondrial functions, and its inactivation may contribute to exercise intolerance and mitochondrial myopathies (Blattler et al., 2012), promoting skeletal muscle wasting. miR-199a-3p affects the eIF4EBP1 gene and reduces the activity of the mTOR pathway, thus interfering with protein synthesis to promote muscle wasting in human skeletal muscle with cancer cachexia (Narasimhan et al., 2017). What’s more, in a meta-analysis, miR-199a is identified as a potential miRNA that inhibits Junb and Caveolin 1 expression, thus suppressing muscle wasting in cancer cachexia (Freire et al., 2019).
Additionally, miRNAs may contribute to muscle wasting via other pathways (Figure 1 and Tables 1, 2). Elevated miR-542-5p may cause muscle atrophy in ICU patients by activating SMAD2/3 phosphorylation. miR-542-5p increases the p-SMAD2/3 level by suppressing the inhibitory components of the TGF-β signaling pathway via a reduction in the expression of SMAD7 and SMURF1, inhibitors of TGF-β type I receptors, and by reducing the phosphatases (such as PPP2CA) that limit TGF-β signaling by dephosphorylating and inactivating SMADs (Garros et al., 2017). In other research, miR-542-3p suppressed ribosomal protein S23 expression and maximal protein synthesis in vitro. In COPD patients, miR-542-3p expression in quadriceps was elevated, and this may suppress physical performance, at least in part, by inhibiting mitochondrial and cytoplasmic ribosome synthesis and suppressing protein synthesis, leading to speculation that miR-542-3p may promote wasting (Farre-Garros et al., 2019). Also, miR-424-5p regulates rRNA synthesis by inhibiting Pol I pre-initiation complex formation, which reduces the capacity of the protein synthesis machinery, contributing to inhibition of protein synthesis and loss of muscle mass in patients with ICU-acquired weakness or COPD, patients undergoing aortic surgery, and in individuals from the Hertfordshire Sarcopenia Study (Connolly et al., 2018). miR-424-5p expression is elevated in muscle of cachectic non-small cell lung cancer (NSCLC) patients compared with healthy control subjects; however, the mechanism underlying the altered expression of miR-424-5p in NSCLC muscle atrophy is unknown (Worp et al., 2020). miR-142a-5p can inhibit mitofusin-1 (MFN1) expression by binding to the 3’UTR of its mRNA. Following overexpression of miR-142a-5p in C2C12 cells and a sciatic nerve transaction mouse model, MFN1 was downregulated, associated with extensive mitochondrial fragmentation, depolarization of mitochondrial membrane potential, accumulation of reactive oxygen species (ROS), and mitophagy and apoptosis activation in skeletal muscle cells, all of which aggravate muscle atrophy. Importantly, these effects were attenuated by overexpression of MFN1 (Yang et al., 2020).
miRNAs That Attenuate Muscle Atrophy
Research strategies based on multiple muscular atrophy models and patient-derived muscle biopsies have also been applied to identify the miRNAs involved in decreasing muscle atrophy in cachexia. These studies have confirmed the crucial roles of miRNAs in regulating muscle wasting in cachexia, and miRNAs in different models of muscle wasting in cachexia can repress wasting via a variety of mechanisms.
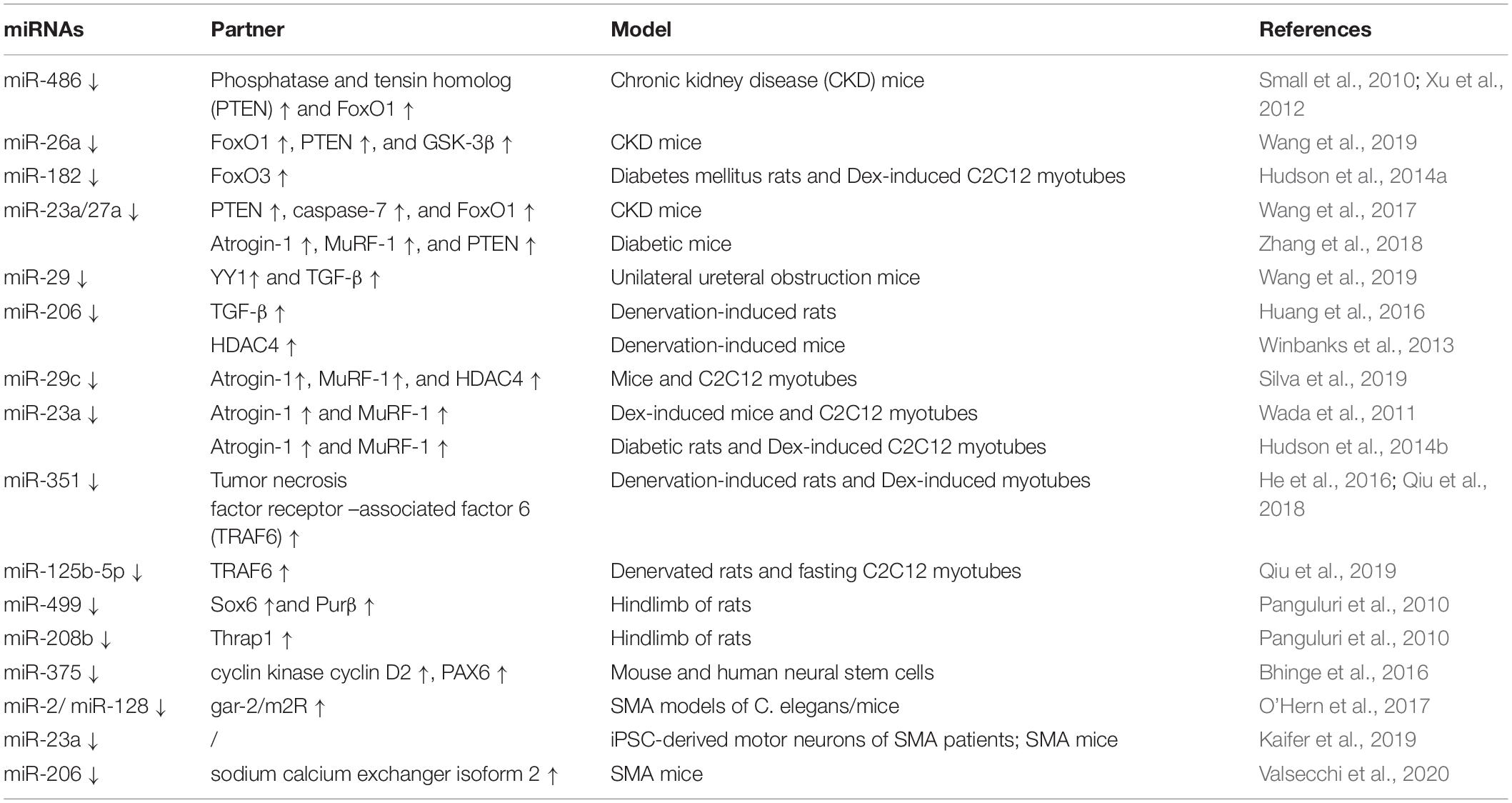
Phosphatase and tensin homolog (PTEN), originally identified as a tumor suppressor, also plays a significant role in skeletal muscle atrophy (Wijesekara et al., 2005; Hu et al., 2007, 2010). It was reported that miRNAs attenuate muscle wasting of cachexia via PTEN/PI3K/Akt signaling (Figure 2 and Table 3). miR-486 and miR-26a repressed the translation of PTEN in a model of CKD, resulting in enhanced phosphorylation of Akt and FoxO1; miR-486 and miR-26a also directly inhibit the translation of FoxO1, and both of these effects lead to suppression of Atrogin-1 and MuRF-1 to prevent muscle atrophy (Small et al., 2010; Xu et al., 2012; Wang B. et al., 2019). miR-182 has also been reported to target FoxO. In muscle cells, miR-182 reduces FoxO3 expression and also blocks glucocorticoid-induced upregulation of multiple FoxO3 targets, including Atrogin-1, autophagy-related protein 12, cathepsin L, and microtubule-associated protein light chain 3, to decrease muscle wasting. miR-182 expression is decreased in C2C12 myotubes with Dex-induced muscle atrophy and in the gastrocnemius muscle of rats injected with streptozotocin-induced diabetes-related atrophy (Hudson et al., 2014a).
Figure 2. Roles of miRNAs in attenuating cachexia skeletal muscle atrophy. The miRNAs are represented in blue; molecules directly targeted by miRNAs are shown in pink; and other signaling molecules involved in muscle wasting are shown in purple.
Overexpression of miR-23a/miR-27a in CKD mice enhanced grip strength, decreased muscle loss, increased the phosphorylation of Akt and FoxO1, decreased the activation of PTEN and FoxO1, and reduced Atrogin-1 and MuRF-1 protein levels, preventing muscle wasting via PTEN/PI3K/Akt signaling (Wang B. et al., 2017). These results are consistent with those of a streptozotocin-induced diabetic muscle atrophy model, and administration of miR-23a/27a weakened the diabetes-induced decrease in the muscle cross-sectional area and muscle function (Zhang A. et al., 2018). PTEN and caspase-7 are targets of miR-23a, and FoxO1 is a target of miR-27a, as identified by luciferase reporter assays in primary satellite cells (Wang B. et al., 2017). In a meta-analysis, miR-27a was showed as a new potential FoxO1 regulator during muscle wasting in cancer cachexia; miR-27a inhibits the expression of FoxO1, so as to inhibit muscle atrophy (Freire et al., 2019).
miR-23a/27a also participates in the myostatin or TGF-β/SMAD signaling pathways to decrease muscle atrophy, similar to other miRNAs, such as miR-29, miR-422a, and miR-206 (Figure 2 and Tables 2, 3). Administration of miR-23a/miR-27a also reduces myostatin expression and its downstream SMAD 2/3 signaling, diminishes caspase 3/7 activation, and increases the expression of muscle regeneration markers (Wang B. et al., 2017). In unilateral ureteral obstruction-induced muscle atrophy, miR-29 ameliorates atrophy and weakens renal fibrosis by suppressing the YY1 and TGF-β signaling proteins directly. YY1 inhibits muscle satellite cell proliferation, leading to the development of muscle wasting (Wang H. et al., 2019). In male patients with ICU-associated muscle weakness or COPD or who were undergoing aortic surgery, quadriceps expression of miR-422a was positively associated with muscle strength. In muscle cells in vitro, miR-422a targeted and restrained SMAD4 expression, a central component of the canonical TGF-β signaling pathway, and suppressed the luciferase activity induced by TGF-β and bone morphogenetic protein (Bmp). In addition, the preoperative miR-422a level is inversely correlated with muscle mass loss during the first week after surgery (Paul et al., 2018).
In a recent study, basic muscle characteristics, such as relative muscle weight, deteriorated continually during a 2-week period after surgical transection of the sciatic nerve. Injection of miR-206 attenuated the deterioration of muscle morphology and physiology, effectively blocked fibrosis, and suppressed TGF-β1 and HDAC4 expression as evaluated at 2 weeks after denervation. Moreover, miR-206 treatment increased the number of differentiating satellite cells, thereby protecting the denervated muscles from atrophy. Interestingly, the ability of miR-206 to regulate HDAC4 expression and to decrease muscle atrophy weakened after pharmacological blockade of the TGF-β/SMAD3 axis (Huang et al., 2016), suggesting that miR-206 may attenuate muscle atrophy via the TGF-β/SMAD3 axis. Other research on miR-206 showed that miR-206 targeted and inhibited HDAC4, and overexpression or inhibition of miR-206 in the muscles of mice reduced or increased HDAC4 levels, respectively, but did not alter muscle mass or myofiber size. Vector-mediated manipulation of miR-206 activity in follistatin-induced hypertrophy and denervation-induced atrophy models did not alter the gain and loss of muscle mass, respectively; miR-206 repressed the hypertrophy of myogenic cells, but not muscle fibers via inhibition of HDAC4 (Winbanks et al., 2013). These two studies showing that miR-206 decreased muscle atrophy are in contrast to another study reporting that miR-206 promotes wasting (Soares et al., 2014). HDAC4 is also targeted by miR-29c, which decreases muscle atrophy (Silva et al., 2019).
In addition to targeting HDAC4, miR-29c may reduce muscle wasting via other pathways (Figure 2 and Table 3). miR-29c can bind to the 3’UTR of MuRF-1 to inhibit its expression. miR-29c is reported to improve skeletal muscle size and function by stimulating satellite cell proliferation and suppressing atrophy-related genes. Overexpression of miR-29c inhibits the expression of Atrogin-1, MuRF-1, and HDAC4 (Silva et al., 2019). miR-23a can also inhibit the translation of both Atrogin-1 and MuRF-1 in a 3’UTR-dependent manner. Ectopic expression of miR-23a is sufficient to protect muscles from Dex-induced atrophy in vitro and in vivo (Wada et al., 2011). In another study, miR-23a was decreased in the gastrocnemius of rats with acute streptozotocin-induced diabetes, a condition known to increase the expression of Atrogin-1 and MuRF-1 and to cause atrophy; miR-23a also reduced in a Dex-induced atrophy model of C2C12 myotubes. The two atrophy-inducing conditions downregulated miR-23a in muscles by mechanisms involving attenuation of the calcineurin/nuclear factor of activated T cells signaling pathway (Hudson et al., 2014b). miR-351 can inhibit denervation-induced atrophy of tibialis anterior (TA) muscles following sciatic nerve transection, at least partially, via negative regulation of tumor necrosis factor receptor-associated factor 6 (TRAF6), as well as Atrogin-1 and MuRF-1, two signaling molecules downstream of TRAF6 (He et al., 2016). This is consistent with the results of the Dex-induced myotube atrophy model (Qiu et al., 2018). TRAF6 is also directly targeted by miR-125b-5p. The expression of miR-125b-5p is downregulated in both atrophic fasting C2C12 myotubes and denervated TA muscles (Qiu et al., 2019), and our research showed that several miRNAs were downregulated under atrophic fasting/starvation conditions in C2C12 myotubes or mice (Lei et al., 2019). Overexpression of miR-125b-5p protects skeletal muscle samples by targeting TRAF6 via downregulation of Atrogin-1, MuRF-1, and autophagy-lysosome system-related proteins (Qiu et al., 2019).
In addition to the above signaling pathway, some miRNAs can decrease muscle atrophy by regulating other molecules (Figure 2 and Table 3). Hindlimb suspension decreases expression of miR-221, miR-499, and miR-208b, increases the levels of Sox6, Purβ (miR-499 targets), and Thrap1 (miR-208b target). Sox6, Purβ, and Thrap1 can decrease β-MyHC expression in rat soleus muscle during skeletal muscle atrophy (McCarthy et al., 2009), suggesting that miR-221, miR-499, and miR-208b may inhibit muscle wasting. Whereas TNF-like weak inducer of apoptosis (TWEAK)-induced skeletal muscle wasting of C2C12 myotubes caused downregulation of miR-1, miR-133a, miR-133b, miR-206, miR-27, miR-23, miR-93, miR-199, miR-107, and miR-192, TWEAK increased the expression of miR-715, miR-146a, miR-455, miR-322, miR-98, and miR-470 in C2C12 myotubes via an unknown mechanism (Panguluri et al., 2010).
miRNAs also play important roles in reducing the pathology of spinal muscular atrophy (SMA) (Table 3). SMA is an autosomal recessive neuromuscular disease, which is caused by deletions or mutations in the survival motor neuron (SMN) gene (Ross and Kwon, 2019). miR-375 promote spinal motor neurogenesis by targeting the cyclin kinase cyclin D2 and the transcription factor Pax6 in mouse and human neural stem cells. Besides, miR-375 inhibits p53 and protects neurons from apoptosis in response to DNA damage, suggesting that miR-375 may play a protective role in motor neurons (Bhinge et al., 2016). In SMA models of C. elegans/mice, miR-2/miR-128 targets gar-2/m2R mRNA in cholinergic neurons and miR-2/miR-128 is required for neuromuscular junction function. miR-2 binds and inhibits gar-2 mRNA translation, but does not reduce transcript levels; and gar-2 loss ameliorates smn-1 neuromuscular defects, suggesting that miR-2/miR-128 may ameliorate neuromuscular defects (O’Hern et al., 2017). In SMA patients and mice, miR-23a significantly reduces the pathology in SMA mice, including increased motor neuron size, reduced neuromuscular junction pathology, increased muscle fiber area, and extended survival (Kaifer et al., 2019). miR-206 reduces the severity of SMA pathology, slowing down disease progression, improving behavioral performance and increasing survival rate of mice. miRNA-206 upregulation induces a reduction of the predicted target sodium calcium exchanger isoform 2, one of the main regulators of intracellular [Ca2+] and [Na+] (Valsecchi et al., 2020).
These data validate the roles of miRNAs in mediating the effects of anabolic and catabolic factors. Especially, experimentally manipulated miRNA expression can attenuate or even reverse skeletal muscle atrophy during cachexia and other diseases, suggesting the substantial contribution of miRNAs in regulating skeletal muscle mass and functions in muscle wasting.
lncRNAs Involved in Skeletal Muscle Atrophy
lncRNAs are ncRNAs longer than 200 nucleotides. A small group of lncRNAs are reportedly expressed in muscle, and they have emerged as significant regulators that promote or attenuate muscle atrophy in cachexia and other diseases (Figure 3 and Table 4). These lncRNAs include the lncRNAs Chronos, atrophy-related long non-coding RNA-1 (Atrolnc-1), mechanical unloading-induced muscle atrophy-related lncRNA (lncMUMA), and lncIRS1 (TCONS-00086268). Studies on the regulation of muscle atrophy in cachexia by lncRNAs are just in the infant stage, the descriptions of lncRNA Pvt1 and SMN-AS1 in other diseases is helpful to a more comprehensive understanding of the relationship between lncRNA and muscle atrophy.
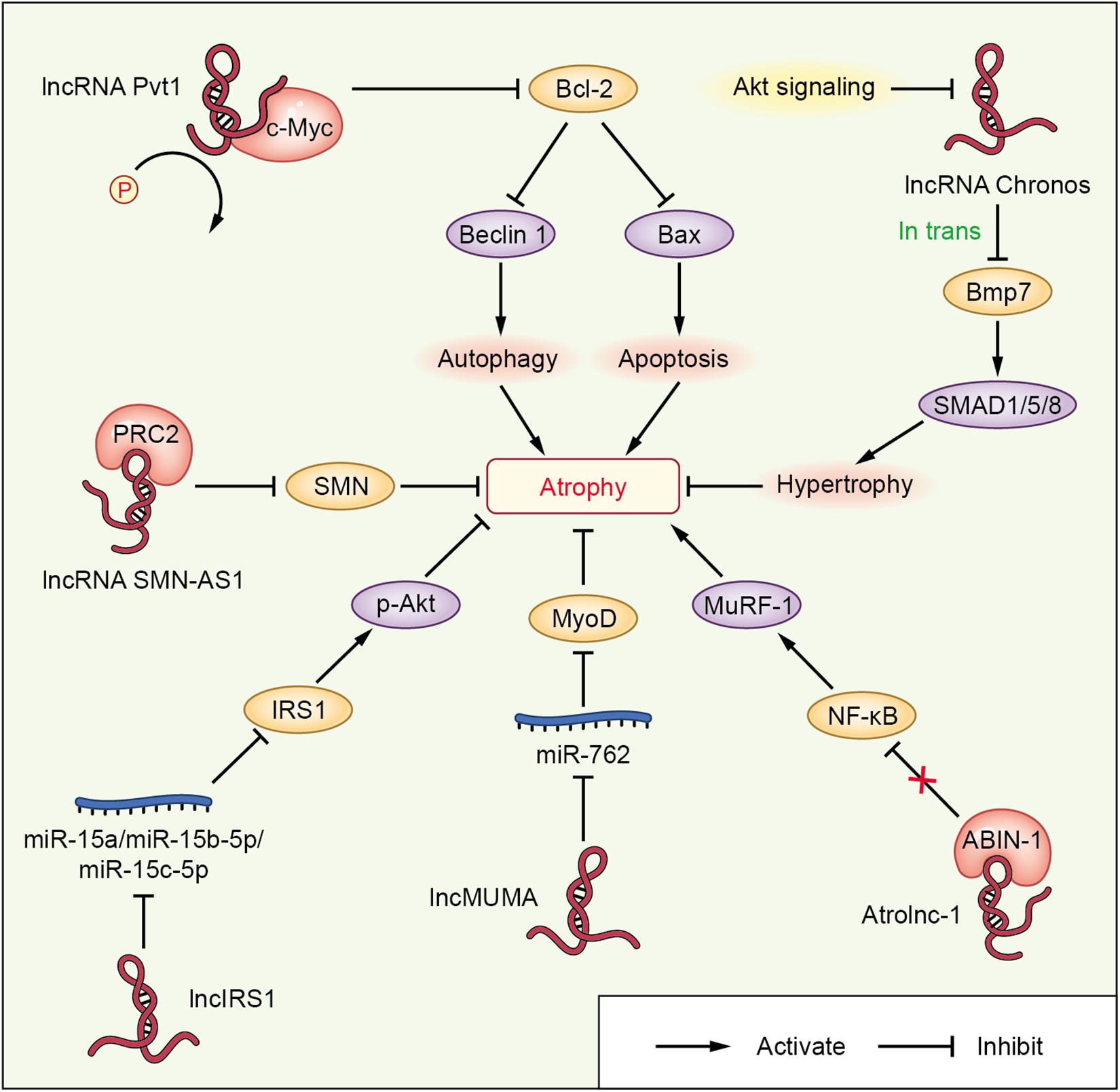
Figure 3. lncRNA-mediated networks regulating skeletal muscle atrophy in cachexia and other diseases. The lncRNAs are represented in red; miRNAs or proteins directly targeted by lncRNAs are shown in blue or pink, respectively; the molecules regulated downstream of miRNAs or proteins interacting with lncRNAs are shown in orange; and other signaling molecules involved in muscle wasting are shown in purple.
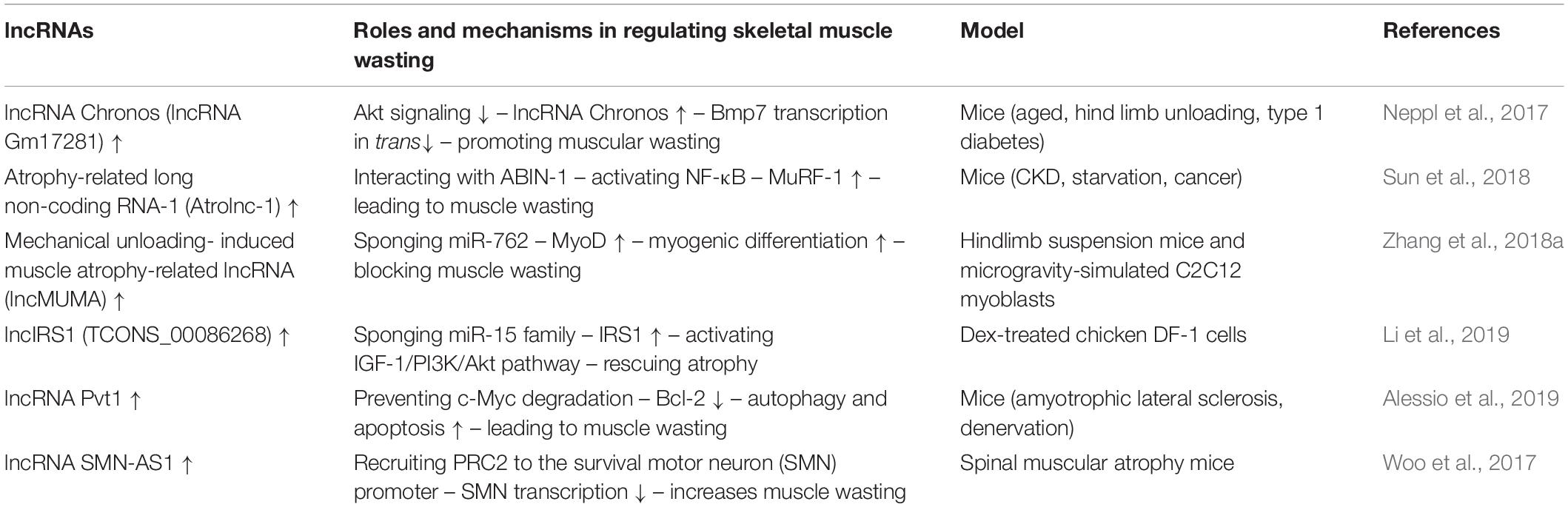
Table 4. lncRNA-mediated networks regulating skeletal muscle atrophy in cachexia and other diseases.
lncRNA Chronos (lncRNA Gm17281), located on chromosome 2, is a muscle-enriched, aging-related lncRNA that is inhibited by Akt and serves as an inhibitor of hypertrophic growth (Neppl et al., 2017), suggesting that Chronos may lead to muscular wasting by restraining hypertrophic growth. Chronos, however, does not appear to function as a classical ‘atrogene’ because its expression is uncoupled from the loss of muscle mass in atrophy models of aging, hind limb unloading, and streptozotocin-induced type 1 diabetes mice. Chronos is negatively regulated by Akt signaling and positively regulated by aging. Chronos can repress hypertrophic growth in vitro and in vivo, in part, by in trans negatively regulating Bmp7 expression, which is located on chromosome 20 (Neppl et al., 2017). This is consistent with prior studies reporting hypertrophic effects of Bmp signaling, which has been shown to positively regulate skeletal muscle hypertrophy via activation of SMAD1/5/8 (Sartori et al., 2013).
Atrolnc-1 increases muscle wasting in fasting mice and mice with cancer, or chronic kidney disease (CKD). Reduced insulin signaling promotes the binding of the transcription factor C/EBP-α to the Atrolnc-1 promoter and increases Atrolnc-1 expression. In cultured C2C12 myotubes, overexpression of Atrolnc-1 promotes protein degradation, whereas Atrolnc-1 knockdown significantly decreases the protein degradation rate increased by serum depletion. Atrolnc-1 interacts with A20 binding inhibitor of NF-κB-1 to inhibit its function, leading to heightened NF-κB activity and MuRF-1 transcription. In the TA muscle of normal mice, overexpression of Atrolnc-1 increases MuRF-1 expression, which results in a loss of myofibers. Knockdown of Atrolnc-1 reduced muscle atrophy in mice with CKD by inhibiting NF-κB activity and MuRF-1 expression (Sun et al., 2018).
In addition to increasing muscle wasting, lncRNAs have also been reported to attenuate muscle atrophy. Mechanical unloading-induced muscle atrophy-related lncRNA (lncMUMA), which is enriched in muscles, can reverse the muscle atrophy induced by hindlimb suspension. lncMUMA is the most downregulated lncRNA during muscle wasting induced by hindlimb suspension mice. In vitro and in vivo data show that the reduction in lncMUMA is closely related to decreased myogenesis during mechanical unloading. In vitro, lncMUMA functions as a miR-762 sponge and facilitates myogenic differentiation by modulating MyoD. Overexpression of lncMUMA alleviates the loss of MyoD protein and muscle mass in miR-762 knock-in mice. In microgravity simulation, overexpression of lncMUMA can accelerate the myogenic differentiation of myoblasts and block the development of muscle wasting in vitro (Zhang et al., 2018a).
lncIRS1 can regulate the expression of atrophy-related genes and rescue muscle wasting. lncIRS1 is particularly enriched in skeletal muscle. lncIRS1 can regulate proliferation and differentiation of myoblasts in vitro and the mass and cross-sectional area of muscle fibers in vivo. The expression of lncIRS1 gradually increases with myogenic differentiation. lncIRS1 can act as a ceRNA for miR-15a, miR-15b-5p, and miR-15c-5p to modulate the expression of IRS1, which is downstream of the IGF-1 receptor. Overexpression of lncIRS1 accelerates the protein expression of IRS1 and facilitates p-Akt signaling, which is a core component of the IGF-1 signaling pathway, leading to the promotion, proliferation, and differentiation of myoblasts and the rescue of muscle atrophy (Li et al., 2019).
Single cell analysis showed that the lncRNA Pvt1, which is activated in early muscle wasting, is involved in promoting muscle atrophy and modulating the mitochondrial network. Pvt1 silencing in models of denervation and amyotrophic lateral sclerosis atrophy resulted in a less fragmented mitochondrial network compared with the controls (Alessio et al., 2019). Pvt1 can prevent c-Myc degradation by hindering phosphorylation of Thr58 in c-Myc (Johnsson and Morris, 2014). c-Myc inhibits the anti-apoptotic protein Bcl-2 (Eischen et al., 2001), which is a key promoter of apoptosis and autophagy, two vital events that occur during muscle atrophy. Bcl-2 can inhibit Beclin 1 and Bax, which enhance autophagy and apoptosis, respectively, leading to muscle wasting (Alessio et al., 2019).
lncRNAs, such as SMN-AS1, also play important roles in accelerating muscle atrophy in SMA. lncRNA SMN-AS1 is transcribed from the antisense strand of the SMN gene and is highly enriched in neurons (D Ydewalle et al., 2017). SMN-AS1 increases muscle wasting by recruiting polycomb repressive complex 2 (PRC2) to the SMN promoter and restricting SMN transcription (Woo et al., 2017). In addition, selective destruction of SMN-AS1-mediated PRC2 recruitment results in activated SMN, attenuated muscle atrophy, and improved SMA phenotypes in mice (Woo et al., 2017).
Recently, several studies have demonstrated the crucial roles of lncRNAs in regulating skeletal muscle atrophy. lncRNAs have been identified as metabolic regulators promoting or reversing muscle atrophy in a variety of wasting models; however, our comprehension of the biological functions of lncRNAs in wasting is still in its infancy. Knowledge of the mechanisms of lncRNA regulation of muscle atrophy may aid the development of potential therapeutics for the clinical treatment of muscle atrophy in cachexia and other diseases.
Conclusion and Perspectives
Skeletal muscle, which can contract or stretch, is vital for body metabolism, homeostasis, and movement (Agudelo et al., 2019; Amoasii et al., 2019; Fiorenza et al., 2019). The development of skeletal muscle has been studied in depth, and a variety of regulatory molecules and complex mechanisms are involved in skeletal muscle development. However, when the normal development of skeletal muscle is disrupted, pathological conditions develop.
Skeletal muscle atrophy in cachexia is due to disruption of the balance between protein synthesis and degradation, which can be partially, but not entirely, reversed by nutritional support (Baracos et al., 2018; de Castro et al., 2019). Weight loss is a significant feature of cachexia and is closely associated with mortality (de Castro et al., 2019). Cachexia is very common in cancer patients, with up to 80% of patients having advanced-stage disease (Porporato, 2016), and cachexia is the immediate cause of death in at least 20% of all cancer patients (Porporato, 2016). Interventions to block or attenuate muscle wasting, while preventing the progression of cachexia, are highly desirable.
The discovery of ncRNAs has opened up a new chapter in medicine (Adams et al., 2017; Rupaimoole and Slack, 2017; Levin, 2019), of which we have just begun to scratch the surface. Focusing on disease-specific and tissue-specific ncRNAs seems to be an ideal approach to identify candidate biomarkers or therapeutic targets, for example, lncRNA PCA3 (also known as DD3) in prostate cancer (Hessels et al., 2003; Tomlins et al., 2011, 2016; Mitra et al., 2020). lncRNA PCA3 is the first ncRNA to receive Food and Drug Administration approval as a cancer biomarker test (Hessels et al., 2003). Although therapeutic applications are still in their infancy, drugs targeting ncRNAs that participate in cancer signaling will hopefully be used to treat cancer patients one day.
In addition to the regulations and potential applications of ncRNAs in cancers, increasing research on the functions of ncRNAs in skeletal muscle suggests that miRNAs and lncRNAs are significant regulators of muscular atrophy in cachexia and other diseases (Crone et al., 2012; Alexander et al., 2014; Narasimhan et al., 2017; Cao et al., 2018; Okugawa et al., 2018; Mitra et al., 2020; Shuai et al., 2020). The roles of miRNAs in skeletal muscle in certain diseases have been well described in different muscular atrophy models, and in recent years, the involvement of miRNAs in regulating muscular atrophy in cachexia has been confirmed. These miRNAs could serve not only as biomarkers for muscle status and wasting but also as biomarkers to monitor muscle regeneration and therapy effects (Suzuki and Springer, 2018). miRNAs associated with muscular atrophy in disease are expected to be identified in the future, and they may function by many different mechanisms. In addition, lncRNAs are reported to participate in various physiological and pathological processes, such as cancer (Lu et al., 2013; He et al., 2014; Sun et al., 2014; Zhuang et al., 2014; Yin et al., 2015; Zhou et al., 2015; Zou et al., 2016; Wang J. et al., 2017), aging (Boon et al., 2016; Su et al., 2019), muscle development (Ballarino et al., 2018; Jin et al., 2018; Zhang et al., 2018b), and their associated disorders (Han et al., 2020). A better understanding of the roles of miRNAs and lncRNAs and their corresponding signaling pathways in regulating skeletal muscle atrophy in cachexia and other diseases will eventually help elucidate the mechanisms underlying muscle wasting and may contribute to the design of new strategies for the prevention, diagnosis, and treatment of muscle-wasting diseases.
In summary, there have been significant advances in our understanding of the crucial roles of miRNAs and lncRNAs in regulating skeletal muscle development (Cesana et al., 2011; Legnini et al., 2014; Zhu et al., 2017; Kong et al., 2019; Cheng et al., 2020; Li et al., 2020). A limited number of studies have specifically identified roles of miRNAs and lncRNAs in muscular atrophy in cachexia and other diseases, and these studies suggest that miRNAs and lncRNAs are central to abnormal regulation of protein synthesis and degradation. Moreover, preliminary evidence suggests that miRNAs and lncRNAs are involved in muscle-organ crosstalk that leads to muscle disorder in COPD (Paul et al., 2018), CKD (Wang B. et al., 2019; Zhang et al., 2019), and heart failure (Kumarswamy et al., 2014; Santer et al., 2019). The roles of altered miRNA (such as miR-29b) and lncRNA levels warrant further investigation in muscle wasting and cachexia (Bei and Xiao, 2017). This phenomenon may hold great potential for the development of possible strategies to treat muscle, pulmonary, kidney, and heart diseases contemporaneously. The challenge is how to use these miRNAs and lncRNAs to develop more effective, economic, preventive, diagnostic, and therapeutic tools to improve the quality of life and decrease the mortality of patients with muscular atrophy in cachexia and other diseases.
Author Contributions
RC conceived the study. RC and SL drafted and revised the manuscript. TJ, YS, and HS performed the literature search and contributed to the draft. All authors read and approved the final version.
Funding
This work was supported by grants from the Natural Science Foundation of Guangdong Province (no. 2018A030313591), the Science and Technology Planning Project of Guangdong Province (no. 2017ZC0333), the Administration of Traditional Chinese Medicine of Guangdong Province (no. 20181014), and the Science and Technology Planning Project of Haizhu District (no. 2018-87).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, B. D., Parsons, C., Walker, L., Zhang, W. C., and Slack, F. J. (2017). Targeting noncoding RNAs in disease. J. Clin. Invest. 127, 761–771. doi: 10.1172/JCI84424
Agudelo, L. Z., Ferreira, D. M. S., Dadvar, S., Cervenka, I., Ketscher, L., Izadi, M., et al. (2019). Skeletal muscle PGC-1α1 reroutes kynurenine metabolism to increase energy efficiency and fatigue-resistance. Nat. Commun. 10:10712. doi: 10.1038/s41467-019-10712-0
Alessio, E., Buson, L., Chemello, F., Peggion, C., Grespi, F., Martini, P., et al. (2019). Single cell analysis reveals the involvement of the long non-coding RNA Pvt1 in the modulation of muscle atrophy and mitochondrial network. Nucleic Acids Res. 47, 1653–1670. doi: 10.1093/nar/gkz007
Alexander, M. S., Casar, J. C., Motohashi, N., Vieira, N. M., Eisenberg, I., Marshall, J. L., et al. (2014). MicroRNA-486–dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy–associated symptoms. J. Clin. Invest. 124, 2651–2667. doi: 10.1172/JCI73579
Amoasii, L., Sanchez-Ortiz, E., Fujikawa, T., Elmquist, J. K., Bassel-Duby, R., and Olson, E. N. (2019). NURR1 activation in skeletal muscle controls systemic energy homeostasis. Proc. Natl. Acad. Sci. U.S.A. 116, 11299–11308. doi: 10.1073/pnas.1902490116
Anker, S. D., Ponikowski, P., Varney, S., Chua, T. P., Clark, A. L., Webb-Peploe, K. M., et al. (1997). Wasting as independent risk factor for mortality in chronic heart failure. Lancet 349, 1050–1053. doi: 10.1016/S0140-6736(96)07015-8
Ballarino, M., Cipriano, A., Tita, R., Santini, T., Desideri, F., Morlando, M., et al. (2018). Deficiency in the nuclear long noncoding RNA Charme causes myogenic defects and heart remodeling in mice. Embo J. 37:e99697. doi: 10.15252/embj.201899697
Baracos, V. E., Martin, L., Korc, M., Guttridge, D. C., and Fearon, K. C. H. (2018). Cancer-associated cachexia. Nat. Rev. Dis. Primers 4:105. doi: 10.1038/nrdp.2017.105
Bei, Y., and Xiao, J. (2017). MicroRNAs in muscle wasting and cachexia induced by heart failure. Nat. Rev. Cardiol. 14:566. doi: 10.1038/nrcardio.2017.122
Bhinge, A., Namboori, S. C., Bithell, A., Soldati, C., Buckley, N. J., and Stanton, L. W. (2016). MiR-375 is essential for human spinal motor neuron development and may be involved in motor neuron degeneration. Stem Cells 34, 124–134. doi: 10.1002/stem.2233
Blattler, S. M., Verdeguer, F., Liesa, M., Cunningham, J. T., Vogel, R. O., Chim, H., et al. (2012). Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor yin yang 1 in skeletal muscle. Mol. Cell Biol. 32, 3333–3346. doi: 10.1128/MCB.00337-12
Boon, R. A., Hofman, P., Michalik, K. M., Lozano-Vidal, N., Berghäuser, D., Fischer, A., et al. (2016). Long noncoding RNA Meg3 controls endothelial cell aging and function. J. Am. Coll Cardiol. 16, 2589–2591. doi: 10.1016/j.jacc.2016.09.949
Cao, L., Liu, Y., Wang, D., Huang, L., Li, F., Liu, J., et al. (2018). MiR-760 suppresses human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1 signaling. J. Exp. Clin. Canc. Res. 37:83. doi: 10.1186/s13046-018-0757-8
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028
Chen, J., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233. doi: 10.1038/ng1725
Chen, J., Tao, Y., Li, J., Deng, Z., Yan, Z., Xiao, X., et al. (2010). microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 190, 867–879. doi: 10.1083/jcb.200911036
Chen, R., Jiang, T., She, Y., Xie, S., Zhou, S., Li, C., et al. (2018). Comprehensive analysis of lncRNAs and mRNAs with associated co-expression and ceRNA networks in C2C12 myoblasts and myotubes. Gene 647, 164–173. doi: 10.1016/j.gene.2018.01.039
Chen, R., Jiang, T., She, Y., Xu, J., Li, C., Zhou, S., et al. (2017). Effects of cobalt chloride, a hypoxia-mimetic agent, on autophagy and atrophy in skeletal C2C12 myotubes. Biomed. Res. Int. 2017, 1–9. doi: 10.1155/2017/7097580
Chen, R., Lei, S., Jiang, T., Zeng, J., Zhou, S., and She, Y. (2020). Roles of lncRNAs and circRNAs in regulating skeletal muscle development. Acta Physiol. (Oxf.) 228:e13356. doi: 10.1111/apha.13356
Chen, R., She, Y., Fu, Q., Chen, X., Shi, H., Lei, S., et al. (2019). Differentially expressed coding and noncoding RNAs in CoCl2–induced cytotoxicity of C2C12 cells. Epigenomics UK 11, 423–438. doi: 10.2217/epi-2018-0087
Cheng, X., Li, L., Shi, G., Chen, L., Fang, C., Li, M., et al. (2020). MEG3 promotes differentiation of porcine satellite cells by sponging miR-423-5p to relieve inhibiting effect on SRF. Cells Basel 9:449. doi: 10.3390/cells9020449
Cichewicz, M. A., Kiran, M., Przanowska, R. K., Sobierajska, E., Shibata, Y., and Dutta, A. (2018). MUNC, an enhancer RNA upstream from the MYOD gene, induces a subgroup of myogenic transcripts in trans independently of MyoD. Mol. Cell Biol. 38:e00655-17. doi: 10.1128/MCB.00655-17
Connolly, M., Paul, R., Farre-Garros, R., Natanek, S. A., Bloch, S., Lee, J., et al. (2018). miR-424-5p reduces ribosomal RNA and protein synthesis in muscle wasting. J. Cachex. Sarcop. Muscle 9, 400–416. doi: 10.1002/jcsm.12266
Crone, S. G., Jacobsen, A., Federspiel, B., Bardram, L., Krogh, A., Lund, A. H., et al. (2012). microRNA-146a inhibits G protein-coupled receptor-mediated activation of NF-κB by targeting CARD10 and COPS8 in gastric cancer. Mol. Cancer 11:71. doi: 10.1186/1476-4598-11-71
de Castro, G. S., Simoes, E., Lima, J. D. C. C., Ortiz-Silva, M., Festuccia, W. T., Tokeshi, F., et al. (2019). Human cachexia induces changes in mitochondria, autophagy and apoptosis in the skeletal muscle. Cancers 11:1264. doi: 10.3390/cancers11091264
Eischen, C. M., Packham, G., Nip, J., Fee, B. E., Hiebert, S. W., Zambetti, G. P., et al. (2001). Bcl-2 is an apoptotic target suppressed by both c-Myc and E2F-1. Oncogene 20, 6983–6993. doi: 10.1038/sj.onc.1204892
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
Farre-Garros, R., Lee, J. Y., Natanek, S. A., Connolly, M., Sayer, A. A., Patel, H., et al. (2019). Quadriceps miR-542-3p and -5p are elevated in COPD and reduce function by inhibiting ribosomal and protein synthesis. J. Appl. Physiol. 126, 1514–1524. doi: 10.1152/japplphysiol.00882.2018
Fearon, K., Strasser, F., Anker, S. D., Bosaeus, I., Bruera, E., Fainsinger, R. L., et al. (2011). Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 12, 489–495. doi: 10.1016/S1470-2045(10)70218-7
Fiorenza, M., Lemminger, A. K., Marker, M., Eibye, K., Marcello Iaia, F., Bangsbo, J., et al. (2019). High-intensity exercise training enhances mitochondrial oxidative phosphorylation efficiency in a temperature–dependent manner in human skeletal muscle: implications for exercise performance. FASEB J. 33, 8976–8989. doi: 10.1096/fj.201900106RRR
Freire, P. P., Fernandez, G. J., Cury, S. S., de Moraes, D., Oliveira, J. S., de Oliveira, G., et al. (2019). The pathway to cancer cachexia: microRNA-regulated networks in muscle wasting based on integrative meta-analysis. Int. J. Mol. Sci. 20:1962. doi: 10.3390/ijms20081962
Garros, R. F., Paul, R., Connolly, M., Lewis, A., Garfield, B. E., Natanek, S. A., et al. (2017). MicroRNA-542 promotes mitochondrial dysfunction and SMAD activity and is elevated in intensive care unit–acquired weakness. Am. J. Resp. Crit. Care 196, 1422–1433. doi: 10.1164/rccm.201701-0101OC
Han, Y., Liu, Y., Yang, C., Gao, C., Guo, X., and Cheng, J. (2020). LncRNA CASC2 inhibits hypoxia-induced pulmonary artery smooth muscle cell proliferation and migration by regulating the miR-222/ING5 axis. Cell Mol. Biol. Lett. 25:215. doi: 10.1186/s11658-020-00215-y
He, Q., Qiu, J., Dai, M., Fang, Q., Sun, X., Gong, Y., et al. (2016). MicroRNA-351 inhibits denervation-induced muscle atrophy by targeting TRAF6. Exp. Ther. Med. 12, 4029–4034. doi: 10.3892/etm.2016.3856
He, Y., Wu, Y., Huang, C., Meng, X., Ma, T., Wu, B., et al. (2014). Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 1842, 2204–2215. doi: 10.1016/j.bbadis.2014.08.015
Hessels, D., Klein Gunnewiek, J. M. T., van Oort, I., Karthaus, H. F. M., van Leenders, G. J. L., van Balken, B., et al. (2003). DD3PCA3-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 44, 8–16. doi: 10.1016/S0302-2838(03)00201-X
Hu, Z., Lee, I. H., Wang, X., Sheng, H., Zhang, L., Du, J., et al. (2007). PTEN expression contributes to the regulation of muscle protein degradation in diabetes. Diabetes 56, 2449–2456. doi: 10.2337/db06-1731
Hu, Z., Wang, H., Lee, I. H., Modi, S., Wang, X., Du, J., et al. (2010). PTEN inhibition improves muscle regeneration in mice fed a high-fat diet. Diabetes 59, 1312–1320. doi: 10.2337/db09-1155
Huang, Q. K., Qiao, H., Fu, M., Li, G., Li, W., Chen, Z., et al. (2016). MiR-206 attenuates denervation-induced skeletal muscle atrophy in rats through regulation of satellite cell differentiation via TGF-β1, Smad3, and HDAC4 signaling. Med. Sci. Monitor. 22, 1161–1170. doi: 10.12659/MSM.897909
Hudson, M. B., Rahnert, J. A., Zheng, B., Woodworth-Hobbs, M. E., Franch, H. A., and Price, S. R. (2014a). miR-182 attenuates atrophy-related gene expression by targeting FoxO3 in skeletal muscle. Am. J. Physiol. Cell Physiol. 307, C314–C319. doi: 10.1152/ajpcell.00395.2013
Hudson, M. B., Woodworth-Hobbs, M. E., Zheng, B., Rahnert, J. A., Blount, M. A., Gooch, J. L., et al. (2014b). miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am. J. Physiol. Cell Physiol. 306, C551–C558. doi: 10.1152/ajpcell.00266.2013
Iannone, F., Montesanto, A., Cione, E., Crocco, P., Caroleo, M. C., Dato, S., et al. (2020). Expression patterns of muscle-specific miR-133b and miR-206 correlate with nutritional status and sarcopenia. Nutrients 12:297. doi: 10.3390/nu12020297
Jin, J. J., Lv, W., Xia, P., Xu, Z. Y., Zheng, A. D., Wang, X. J., et al. (2018). Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. U.S.A. 115, E9802–E9811. doi: 10.1073/pnas.1801471115
Johnsson, P., and Morris, K. V. (2014). Expanding the functional role of long noncoding RNAs. Cell Res. 24, 1284–1285. doi: 10.1038/cr.2014.104
Kaifer, K. A., Villalón, E., O’Brien, B. S., Sison, S. L., Smith, C. E., Simon, M. E., et al. (2019). AAV9-mediated delivery of miR-23a reduces disease severity in Smn2B/–SMA model mice. Hum. Mol. Genet. 28, 3199–3210. doi: 10.1093/hmg/ddz142
Kong, D., He, M., Yang, L., Zhou, R., Yan, Y. Q., Liang, Y., et al. (2019). MiR-17 and miR-19 cooperatively promote skeletal muscle cell differentiation. Cell Mol. Life Sci. 76, 5041–5054. doi: 10.1007/s00018-019-03165-7
Kukreti, H., Amuthavalli, K., Harikumar, A., Sathiyamoorthy, S., Feng, P. Z., Anantharaj, R., et al. (2013). Muscle-specific MicroRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J. Biol. Chem. 288, 6663–6678. doi: 10.1074/jbc.M112.390369
Kumarswamy, R., Bauters, C., Volkmann, I., Maury, F., Fetisch, J., Holzmann, A., et al. (2014). Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 114, 1569–1575. doi: 10.1161/CIRCRESAHA.114.303915
Legnini, I., Morlando, M., Mangiavacchi, A., Fatica, A., and Bozzoni, I. (2014). A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell 53, 506–514. doi: 10.1016/j.molcel.2013.12.012
Lei, S., She, Y., Zeng, J., Chen, R., Zhou, S., and Shi, H. (2019). Expression patterns of regulatory lncRNAs and miRNAs in muscular atrophy models induced by starvation in vitro and in vivo. Mol. Med. Rep. 20, 4175–4185. doi: 10.3892/mmr.2019.10661
Levin, A. A. (2019). Treating disease at the RNA level with oligonucleotides. New Engl. J. Med. 380, 57–70. doi: 10.1056/NEJMra1705346
Li, J., Chan, M. C., Yu, Y., Bei, Y., Chen, P., Zhou, Q., et al. (2017). miR-29b contributes to multiple types of muscle atrophy. Nat. Commun. 8:15201. doi: 10.1038/ncomms15201
Li, J., Wang, L., Hua, X., Tang, H., Chen, R., Yang, T., et al. (2020). CRISPR/Cas9-mediated miR-29b editing as a treatment of different types of muscle atrophy in mice. Mol. Ther. 28:5. doi: 10.1016/j.ymthe.2020.03.005
Li, Z., Cai, B., Abdalla, B. A., Zhu, X., Zheng, M., Han, P., et al. (2019). LncIRS1 controls muscle atrophy via sponging miR - 15 family to activate IGF1–PI3K/AKT pathway. J. Cachex. Sarcop. Muscle 10, 391–410. doi: 10.1002/jcsm.12374
Liu, C., Wang, M., Chen, M., Zhang, K., Gu, L., Li, Q., et al. (2017). miR-18a induces myotubes atrophy by down-regulating IgfI. Int. J. Biochem. Cell Biol. 90, 145–154. doi: 10.1016/j.biocel.2017.07.020
Lu, K., Li, W., Liu, X., Sun, M., Zhang, M., Wu, W., et al. (2013). Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 13:461. doi: 10.1186/1471-2407-13-461
Lu, L., Sun, K., Chen, X., Zhao, Y., Wang, L., Zhou, L., et al. (2013). Genome–wide survey by ChIP - seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J. 32, 2575–2588. doi: 10.1038/emboj.2013.182
Man, W. D., Kemp, P., Moxham, J., and Polkey, M. I. (2009). Skeletal muscle dysfunction in COPD: clinical and laboratory observations. Clin. Sci. (Lond.) 117, 251–264. doi: 10.1042/CS20080659
McCarthy, J. J., Esser, K. A., Peterson, C. A., and Dupont-Versteegden, E. E. (2009). Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol. Genomics 39, 219–226. doi: 10.1152/physiolgenomics.00042.2009
Mitra, R., Adams, C. M., Jiang, W., Greenawalt, E., and Eischen, C. M. (2020). Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival. Nat. Commun. 11:968. doi: 10.1038/s41467-020-14713-2
Narasimhan, A., Ghosh, S., Stretch, C., Greiner, R., Bathe, O. F., Baracos, V., et al. (2017). Small RNAome profiling from human skeletal muscle: novel miRNAs and their targets associated with cancer cachexia. J. Cachex. Sarcop. Muscle 8, 405–416. doi: 10.1002/jcsm.12168
Neppl, R. L., Wu, C. L., and Walsh, K. (2017). lncRNA Chronos is an aging-induced inhibitor of muscle hypertrophy. J. Cell Biol. 216, 3497–3507. doi: 10.1083/jcb.201612100
O’Hern, P. J., Do Carmo, G., Gonçalves, I., Brecht, J., López Soto, E. J., Simon, J., et al. (2017). Decreased microRNA levels lead to deleterious increases in neuronal M2 muscarinic receptors in Spinal Muscular Atrophy models. Elife 6:20752. doi: 10.7554/eLife.20752
Okugawa, Y., Yao, L., Toiyama, Y., Yamamoto, A., Shigemori, T., Yin, C., et al. (2018). Prognostic impact of sarcopenia and its correlation with circulating miR-21 in colorectal cancer patients. Oncol. Rep. 39, 1555–1564. doi: 10.3892/or.2018.6270
Panguluri, S. K., Bhatnagar, S., Kumar, A., McCarthy, J. J., Srivastava, A. K., Cooper, N. G., et al. (2010). Genomic profiling of messenger RNAs and microRNAs reveals potential mechanisms of TWEAK-induced skeletal muscle wasting in mice. PLoS One 5:e8760. doi: 10.1371/journal.pone.0008760
Paul, R., Lee, J., Donaldson, A. V., Connolly, M., Sharif, M., Natanek, S. A., et al. (2018). miR-422a suppresses SMAD4 protein expression and promotes resistance to muscle loss. J. Cachexia. Sarcopenia. Muscle 9, 119–128. doi: 10.1002/jcsm.12236
Ponnusamy, M., Liu, F., Zhang, Y., Li, R., Zhai, M., Liu, F., et al. (2019). Long noncoding RNA CPR (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation 139, 2668–2684. doi: 10.1161/CIRCULATIONAHA.118.035832
Porporato, P. E. (2016). Understanding cachexia as a cancer metabolism syndrome. Oncogenesis 5:e200. doi: 10.1038/oncsis.2016.3
Prado, C. M., Lieffers, J. R., McCargar, L. J., Reiman, T., Sawyer, M. B., Martin, L., et al. (2008). Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: a population-based study. Lancet Oncol. 9, 629–635. doi: 10.1016/S1470-2045(08)70153-0
Qiu, J., Wang, L., Wang, Y., Zhang, Q., Ma, W., Fang, Q., et al. (2018). MicroRNA351 targeting TRAF6 alleviates dexamethasone-induced myotube atrophy. J. Thorac. Dis. 10, 6238–6246. doi: 10.21037/jtd.2018.10.88
Qiu, J., Zhu, J., Zhang, R., Liang, W., Ma, W., Zhang, Q., et al. (2019). miR-125b-5p targeting TRAF6 relieves skeletal muscle atrophy induced by fasting or denervation. Ann. Transl. Med. 7:456. doi: 10.21037/atm.2019.08.39
Ramalho-Carvalho, J., Martins, J. B., Cekaite, L., Sveen, A., Torres-Ferreira, J., Graca, I., et al. (2017). Epigenetic disruption of miR-130a promotes prostate cancer by targeting SEC23B and DEPDC1. Cancer Lett. 385, 150–159. doi: 10.1016/j.canlet.2016.10.028
Rom, O., and Reznick, A. Z. (2016). The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Radical. Bio. Med. 98, 218–230. doi: 10.1016/j.freeradbiomed.2015.12.031
Ross, L. F., and Kwon, J. M. (2019). Spinal muscular atrophy: past, present, and future. Neoreviews 20, e437–e451. doi: 10.1542/neo.20-8-e437
Rupaimoole, R., and Slack, F. J. (2017). MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222. doi: 10.1038/nrd.2016.246
Santer, L., López, B., Ravassa, S., Baer, C., Riedel, I., Chatterjee, S., et al. (2019). Circulating long noncoding RNA LIPCAR predicts heart failure outcomes in patients without chronic kidney disease. Hypertension 73, 820–828. doi: 10.1161/HYPERTENSIONAHA.118.12261
Sartori, R., Schirwis, E., Blaauw, B., Bortolanza, S., Zhao, J., Enzo, E., et al. (2013). BMP signaling controls muscle mass. Nat. Genet. 45, 1309–1318. doi: 10.1038/ng.2772
Shuai, Y., Ma, Z., Liu, W., Yu, T., Yan, C., Jiang, H., et al. (2020). TEAD4 modulated LncRNA MNX1-AS1 contributes to gastric cancer progression partly through suppressing BTG2 and activating BCL2. Mol. Cancer 19:6. doi: 10.1186/s12943-019-1104-1
Silva, W. J., Graça, F. A., Cruz, A., Silvestre, J. G., Labeit, S., Miyabara, E. H., et al. (2019). miR–29c improves skeletal muscle mass and function throughout myocyte proliferation and differentiation and by repressing atrophy - related genes. Acta Physiol. 226:13278. doi: 10.1111/apha.13278
Small, E. M., O’Rourke, J. R., Moresi, V., Sutherland, L. B., McAnally, J., Gerard, R. D., et al. (2010). Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U.S.A. 107, 4218–4223. doi: 10.1073/pnas.1000300107
Soares, R. J., Cagnin, S., Chemello, F., Silvestrin, M., Musaro, A., De Pitta, C., et al. (2014). Involvement of MicroRNAs in the regulation of muscle wasting during catabolic conditions. J. Biol. Chem. 289, 21909–21925. doi: 10.1074/jbc.M114.561845
Sonkoly, E., Bata-Csorgo, Z., Pivarcsi, A., Polyanka, H., Kenderessy-Szabo, A., Molnar, G., et al. (2005). Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J. Biol. Chem. 280, 24159–24167. doi: 10.1074/jbc.M501704200
Su, Z., Xiong, H., Pang, J., Lin, H., Lai, L., Zhang, H., et al. (2019). LncRNA AW112010 promotes mitochondrial biogenesis and hair cell survival: implications for age-related hearing loss. Oxid. Med. Cell Longev. 2019, 1–13. doi: 10.1155/2019/6150148
Sun, L., Si, M., Liu, X., Choi, J. M., Wang, Y., Thomas, S. S., et al. (2018). Long-noncoding RNA Atrolnc-1 promotes muscle wasting in mice with chronic kidney disease. J. Cachex. Sarcopenia Muscle 9, 962–974. doi: 10.1002/jcsm.12321
Sun, M., Xia, R., Jin, F., Xu, T., Liu, Z., De, W., et al. (2014). Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumor Biol. 35, 1065–1073. doi: 10.1007/s13277-013-1142-z
Suzuki, T., and Springer, J. (2018). MicroRNAs in muscle wasting. J. Cachex. Sarcopenia Muscle 9, 1209–1212. doi: 10.1002/jcsm.12384
Tomlins, S. A., Aubin, S. M., Siddiqui, J., Lonigro, R. J., Sefton-Miller, L., Miick, S., et al. (2011). Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci. Transl. Med. 3, 72r–94r. doi: 10.1126/scitranslmed.3001970
Tomlins, S. A., Day, J. R., Lonigro, R. J., Hovelson, D. H., Siddiqui, J., Kunju, L. P., et al. (2016). Urine TMPRSS2:ERG plus PCA3 for individualized prostate cancer risk assessment. Eur. Urol. 70, 45–53. doi: 10.1016/j.eururo.2015.04.039
Valsecchi, V., Anzilotti, S., Serani, A., Laudati, G., Brancaccio, P., Guida, N., et al. (2020). miR-206 reduces the severity of motor neuron degeneration in the facial nuclei of the brainstem in a mouse model of SMA. Mol. Ther. 28, 1154–1166. doi: 10.1016/j.ymthe.2020.01.013
Wada, S., Kato, Y., Okutsu, M., Miyaki, S., Suzuki, K., Yan, Z., et al. (2011). Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J. Biol. Chem. 286, 38456–38465. doi: 10.1074/jbc.M111.271270
Wang, B., Zhang, A., Wang, H., Klein, J. D., Tan, L., Wang, Z., et al. (2019). miR-26a limits muscle wasting and cardiac fibrosis through exosome-mediated microRNA transfer in chronic kidney disease. Theranostics 9, 1864–1877. doi: 10.7150/thno.29579
Wang, B., Zhang, C., Zhang, A., Cai, H., Price, S. R., and Wang, X. H. (2017). MicroRNA-23a and MicroRNA-27a mimic exercise by ameliorating CKD-induced muscle atrophy. J. Am. Soc. Nephrol. 28, 2631–2640. doi: 10.1681/ASN.2016111213
Wang, H., Wang, B., Zhang, A., Hassounah, F., Seow, Y., Wood, M., et al. (2019). Exosome-mediated miR-29 transfer reduces muscle atrophy and kidney fibrosis in mice. Mol. Ther. 27, 571–583. doi: 10.1016/j.ymthe.2019.01.008
Wang, J., Cao, B., Han, D., Sun, M., and Feng, J. (2017). Long Non-coding RNA H19 induces cerebral ischemia reperfusion injury via activation of autophagy. Aging Dis. 8:71. doi: 10.14336/AD.2016.0530
Wijesekara, N., Konrad, D., Eweida, M., Jefferies, C., Liadis, N., Giacca, A., et al. (2005). Muscle-specific pten deletion protects against insulin resistance and diabetes. Mol. Cell Biol. 25, 1135–1145. doi: 10.1128/MCB.25.3.1135-1145.2005
Winbanks, C. E., Beyer, C., Hagg, A., Qian, H., Sepulveda, P. V., and Gregorevic, P. (2013). miR-206 represses hypertrophy of myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS One 8:e73589. doi: 10.1371/journal.pone.0073589
Woo, C. J., Maier, V. K., Davey, R., Brennan, J., Li, G., Brothers, J., et al. (2017). Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 114, E1509–E1518. doi: 10.1073/pnas.1616521114
Worp, W. R. P. H., Schols, A. M. W. J., Dingemans, A. M. C., Op Den, Kamp, C. M. H., Degens, J. H. R. J., et al. (2020). Identification of microRNAs in skeletal muscle associated with lung cancer cachexia. J. Cachex. Sarcopenia Muscle 11, 452–463. doi: 10.1002/jcsm.12512
Xu, J., Li, R., Workeneh, B., Dong, Y., Wang, X., and Hu, Z. (2012). Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int. 82, 401–411. doi: 10.1038/ki.2012.84
Yang, X., Xue, P., Chen, H., Yuan, M., Kang, Y., Duscher, D., et al. (2020). Denervation drives skeletal muscle atrophy and induces mitochondrial dysfunction, mitophagy and apoptosis via miR-142a-5p/MFN1 axis. Theranostics 10, 1415–1432. doi: 10.7150/thno.40857
Ydewalle, D., Ramos, D. M., Pyles, N. J., Ng, S., Gorz, M., Pilato, C. M., et al. (2017). The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron 93, 66–79. doi: 10.1016/j.neuron.2016.11.033
Yin, D. D., Liu, Z. J., Zhang, E., Kong, R., Zhang, Z. H., and Guo, R. H. (2015). Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. 36, 4851–4859. doi: 10.1007/s13277-015-3139-2
Yu, Y., Li, X., Liu, L., Chai, J., Haijun, Z., Chu, W., et al. (2016). miR-628 promotes burn-induced skeletal muscle atrophy via targeting IRS1. Int. J. Biol. Sci. 12, 1213–1224. doi: 10.7150/ijbs.15496
Zhang, A., Li, M., Wang, B., Klein, J. D., Price, S. R., and Wang, X. H. (2018). miRNA-23a/27a attenuates muscle atrophy and renal fibrosis through muscle-kidney crosstalk. J. Cachex. Sarcopenia Muscle 9, 755–770. doi: 10.1002/jcsm.12296
Zhang, A., Wang, H., Wang, B., Yuan, Y., Klein, J. D., and Wang, X. H. (2019). Exogenous miR-26a suppresses muscle wasting and renal fibrosis in obstructive kidney disease. FASEB J. 33, 13590–13601. doi: 10.1096/fj.201900884R
Zhang, Z. K., Li, J., Guan, D., Liang, C., Zhuo, Z., Liu, J., et al. (2018a). Long noncoding RNA lncMUMA reverses established skeletal muscle atrophy following mechanical unloading. Mol. Ther. 26, 2669–2680. doi: 10.1016/j.ymthe.2018.09.014
Zhang, Z. K., Li, J., Guan, D., Liang, C., Zhuo, Z., Liu, J., et al. (2018b). A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachex. Sarcopenia Muscle 9, 613–626. doi: 10.1002/jcsm.12281
Zhou, X., Yin, C., Dang, Y., Ye, F., and Zhang, G. (2015). Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep.UK 5:11516. doi: 10.1038/srep11516
Zhu, M., Liu, J., Xiao, J., Yang, L., Cai, M., Shen, H., et al. (2017). Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 8:14718. doi: 10.1038/ncomms14718
Zhuang, M., Gao, W., Xu, J., Wang, P., and Shu, Y. (2014). The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Co. 448, 315–322. doi: 10.1016/j.bbrc.2013.12.126
Keywords: skeletal muscle, atrophy, cachexia, miRNAs, lncRNAs
Citation: Chen R, Lei S, Jiang T, She Y and Shi H (2020) Regulation of Skeletal Muscle Atrophy in Cachexia by MicroRNAs and Long Non-coding RNAs. Front. Cell Dev. Biol. 8:577010. doi: 10.3389/fcell.2020.577010
Received: 28 June 2020; Accepted: 26 August 2020;
Published: 15 September 2020.
Edited by:
Jaime J. Carvajal, Andalusian Centre for Development Biology (CABD), SpainReviewed by:
Amelia Eva Aranega, University of Jaén, SpainSheik Pran Babu Sardar Pasha, University of California, Davis, United States
Copyright © 2020 Chen, Lei, Jiang, She and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Chen, rui.c.med@163.com
 Rui Chen
Rui Chen Si Lei1
Si Lei1