- Review
- Open access
- Published:
From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis
Genome Biology volume 21, Article number: 22 (2020)
Abstract
Assay of Transposase Accessible Chromatin sequencing (ATAC-seq) is widely used in studying chromatin biology, but a comprehensive review of the analysis tools has not been completed yet. Here, we discuss the major steps in ATAC-seq data analysis, including pre-analysis (quality check and alignment), core analysis (peak calling), and advanced analysis (peak differential analysis and annotation, motif enrichment, footprinting, and nucleosome position analysis). We also review the reconstruction of transcriptional regulatory networks with multiomics data and highlight the current challenges of each step. Finally, we describe the potential of single-cell ATAC-seq and highlight the necessity of developing ATAC-seq specific analysis tools to obtain biologically meaningful insights.
Introduction
Mammalian DNA is highly condensed through three major hierarchical scales; the first is the nucleosome which then wraps into chromatin leading to the third hierarchy, the chromosome [1,2,3,4,5,6]. Chromatin can dynamically switch between transcriptionally active euchromatin and inactive heterochromatin [7, 8]. All three scales of DNA condensation and their interplay contribute to gene regulation.
Recent gene regulation studies have focused on epigenetics, and through the advances of high-throughput sequencing technologies, various assays have been developed to decipher the epigenetic landscape. These include Assay of Transposase Accessible Chromatin sequencing (ATAC-seq) [9, 10], DNase I hypersensitive sites sequencing (DNase-seq) [11,12,13] and Formaldehyde-Assisted Isolation of Regulatory Elements sequencing (FAIRE-seq) [14], all of which interrogate chromatin accessibility; Chromatin Immuno-Precipitation sequencing (ChIP-seq) which measures transcription factor (TF) binding [15,16,17] and histone modifications [18, 19]; and Micrococcal Nuclease sequencing (MNase-seq) which detects nucleosome positioning and occupancy [20, 21]. Detailed procedures of these assays are out of scope of this review and discussed in detail elsewhere [22].
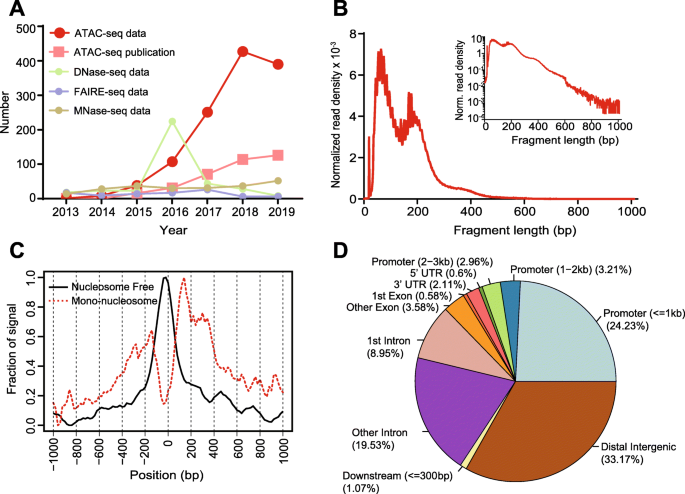
Among assays designed for detecting chromatin accessibility, ATAC-seq has gained particular popularity since first described in 2013. An exponential increase of curated ATAC-seq datasets and publications indicates its value in a wide spectrum of biological questions (Fig. 1a), such as depicting enhancer landscapes in healthy mammalian tissue and cell types [23,24,25], studying accessibility changes between normal hematopoiesis and leukemia [26, 27], as well as the chromatin state within schizophrenia patients and the Cancer Genome Atlas (TCGA) pan-cancer cohort [28, 29]. A schematic diagram of this cutting-edge technology in fundamental and translational research is shown in Fig. 3a. Briefly, ATAC-seq incorporates a genetically engineered hyperactive Tn5 transposase that simultaneously cuts open chromatin leaving a 9-bp staggered nick and ligates high-throughput sequencing adapters to these regions. During this process, the nick is repaired, leaving a 9-bp duplication [30, 31]. Paired-end sequencing is then performed to facilitate higher unique alignment rates of these open regions [32].
Overview of ATAC-seq datasets increase and sample output for pre-analysis and advanced analysis. a The number of ATAC-seq datasets, ATAC-seq publications, DNase-seq datasets, FAIRE-seq datasets, and MNase-seq datasets in PubMed from 1 Jan 2013 to 1 Oct 2019. b Typical fragment size distribution plot shows enrichment around 100 and 200 bp, indicating nucleosome-free and mono-nucleosome-bound fragments. c Typical TSS enrichment plot shows that nucleosome-free fragments are enriched at TSS, while mono-nucleosome fragments are depleted at TSS but enriched at flanking regions. d Typical peak annotation pie chart shows that more than half of the peaks fall into enhancer regions (distal intergenic and intronic regions), and only around 25% of the peaks are in promoter regions. TSS: transcription start site
The hyperactivity of Tn5 transposase makes the ATAC-seq protocol a simple, time-efficient method that requires 500–50,000 cells [9]. The sensitivity and specificity are comparable to DNase-seq but superior to FAIRE-seq where both methods require millions of cells as input material [9]. Because ATAC-seq does not involve rigorous size selection during library preparation, it can also identify nucleosome positions using fragments representing nucleosome monomer and multi-mers [9]. Recently, single-cell ATAC-seq (scATAC-seq) has been described, using fluorescence-activated cell sorting (FACS), microfluidic, and nano-well-based approaches [33,34,35]. ScATAC-seq can be applied in multiple situations including clinical specimens and developmental biology to study the heterogenous cell populations at single-cell resolution [23, 29].
Despite the simplicity and robustness of ATAC-seq, a major impediment exists as there are few bioinformatic analysis tools developed specifically for ATAC-seq data [32, 36]. Analysis tools used in ChIP-seq and DNase-seq have been applied to ATAC-seq assuming similar data characteristics [37]. However, this assumption has not been evaluated systematically.
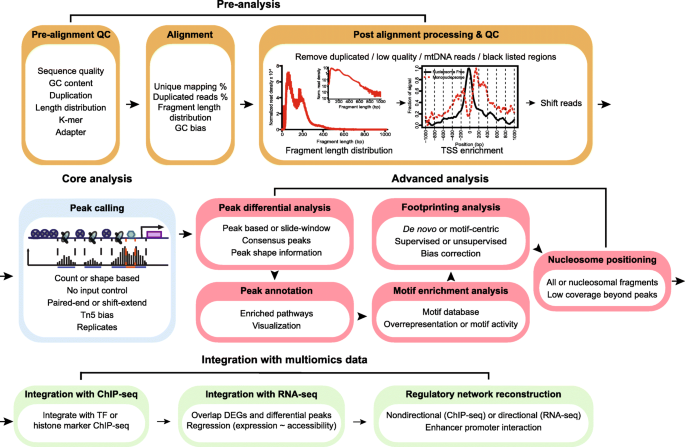
The major focus of this review is to discuss current resources for ATAC-seq analysis. We aim to provide an annotated guide for ATAC-seq data analysis instead of an exhaustive collection of tools. Previous reviews regarding ATAC-seq data analysis have focused mainly on peak callers and modeling regulatory networks [37, 38], but a systematic review covering major parts of ATAC-seq data analysis is urgently needed. This review will cover the four most important steps listed in the flowchart (Fig. 2). These include (1) pre-analysis (quality control (QC) and alignment), (2) core analysis (peak calling), (3) advanced analysis at the level of peaks, motifs, nucleosomes, and TF footprints, and (4) integration with multiomics data to reconstruct regulatory networks. These steps will allow researchers to conduct robust analysis on ATAC-seq data and generate more biological meaningful results. Finally, we will present the challenges and opportunities of ATAC-seq analysis and scATAC-seq.
Roadmap of a typical ATAC-seq analysis. Four major steps are listed, including pre-analysis, core analysis, advanced analysis, and integration with multiomics data. Pre-analyses include pre-alignment QC, alignment and post-alignment processing, and QC. Core analysis includes peak calling. Advanced analyses include peak, motif, footprint, and nucleosome analysis. Multiomics data integration includes integration with ChIP-seq and RNA-seq data and regulatory network reconstruction. Text in each box emphasizes the important considerations in each analysis step. We suggest researchers start with FastQC, trimmomatic, and BWA-MEM for pre-analysis, MACS2 for peak calling, csaw for peak differential analysis, ChIPseeker for annotation and visualization, MEME suite for motif detection and enrichment, HMMRATAC for nucleosome detection, HINT-ATAC for footprint analysis, and PCEA for regulatory network reconstruction with RNA-seq. QC: quality check; TSS: transcription start site; TF: transcription factor; DEG: differentially expressed gene
Pre-analysis: quality control and alignment
The first step of ATAC-seq analysis involves pre-alignment QC, read alignment to a reference genome, and post-alignment QC and processing (Fig. 2a) [32].
Pre-alignment quality control
The pre-alignment QC and read alignment steps are standard for most high-throughput sequencing technologies. For example, FastQC [39] can be used to visualize base quality scores, GC content, sequence length distribution, sequence duplication levels, k-mer overrepresentation and contamination of primers and adapters in the sequencing data. An overall high base quality score with a slight drop towards the 3′ end of sequencing reads is acceptable. No obvious deviation from expected GC content and sequence read length should be observed. Moreover, the metrics should be homogeneous among all samples from the same experimental batch and sequencing run.
Currently, due to the ubiquitous use of Illumina’s Nextera library for ATAC-seq, overrepresentation of Nextera sequencing adapters is often observed and should be removed for accurate read alignment. Most adapter removal tools employ different variations of dynamic programming, such as cutadapt [40], AdapterRemoval v2 [41], Skewer [42], and trimmomatic [43] all requiring input of known adapter sequences. For example, using trimmomatic with built-in adapter sequences for Nextera and TruSeq library would be a straightforward step. Low-quality bases can also be eliminated using these tools. From our experience, read trimming tools are generally comparable in performance of efficient removal of low-quality and contaminating adapter sequences.
Alignment
After read trimming, FastQC can be performed again to check the successful removal of adapter and low-quality bases. Trimmed reads are then mapped to a reference genome. BWA-MEM [44] and Bowtie2 [45] aligners are memory-efficient and fast for short paired-end reads. The soft-clip strategy from both aligners allows the overhang of bases on both ends of reads which can further increase unique mapping rates [46]. We suggest that a unique mapping rate over 80% is typical for a successful ATAC-seq experiment. For mammalian species, the suggested minimum number of mapped reads is 50 million for open chromatin detection and differential analysis, and 200 million for TF footprinting based on empirical and computational estimations [10, 12, 47,48,49].
Post-alignment processing and quality control
After sequence alignment, as in most DNA sequencing data, basic metrics of the aligned BAM file, such as unique mapping reads/rates, duplicated read percentages, and fragment size distribution can be collected using Picard [50] and SAMtools [51]. Additionally, reads should be removed if they are improperly paired or of low mapping quality. The mitochondrial genome, which is more accessible due to the lack of chromatin packaging [52], and the ENCODE blacklisted regions [53, 54] often have extremely high read coverage, and should also be discarded [33]. Duplicated reads, which are likely to have arisen as PCR artifacts, should also be removed to significantly improve biological reproducibility [48]. These steps will together improve the power of open chromatin detection and produce fewer false positives.
There are additional ATAC-seq-specific quality metrics that need to be evaluated. Typically, a successful ATAC-seq experiment should generate a fragment size distribution plot with decreasing and periodical peaks corresponding to the nucleosome-free regions (NFR) (< 100 bp) and mono-, di-, and tri-nucleosomes (~ 200, 400, 600 bp, respectively) (Fig. 1b) [9, 55]. Fragments from the NFR are expected to be enriched around the transcription start site (TSS) of genes, while fragments from nucleosome-bound regions are expected to be depleted at TSS with a slight enrichment of flanking regions around TSS (Fig. 1c) [55]. These can be evaluated with the tool ATACseqQC [55]. Lastly, reads should be shifted + 4 bp and − 5 bp for positive and negative strand respectively, to account for the 9-bp duplication created by DNA repair of the nick by Tn5 transposase and achieve base-pair resolution of TF footprint and motif-related analyses [9, 33, 56]. Most aforementioned QC and analysis reports can be integrated using MultiQC [57] for an aggregated, user-friendly, and interactive presentation.
A major consideration for the appropriate tools to choose here is often time to result. Read trimming and alignment can be time consuming, and there is always a trade-off between speed and accuracy. In our experience, the following pipeline performs reasonably well: FastQC➔ trimmomatic➔BWA-MEM➔ATACseqQC, and we would suggest this as a good starting point for processing of ATAC-seq data.
Core analysis: peak calling
The second major step of ATAC-seq data analysis is to identify accessible regions (also referred to as peaks) and is the basis for advanced analysis. A similar process has been comprehensively reviewed for ChIP-seq [58, 59] and DNase-seq [60]. Currently, MACS2 is the default peak caller of the ENCODE ATAC-seq pipeline. To the best of our knowledge, only one peak caller is specifically developed for ATAC-seq [61]. All others are adopted from ChIP-seq and DNase-seq with the assumption that ATAC-seq peak patterns share the same properties. Thus, we will focus on tools that are currently used in ATAC-seq and provide an overview of potential alternatives (Fig. 4a).
Unlike in ChIP-seq, input controls for ATAC-seq, in which Tn5 transposase randomly cleaves protein-free DNA, are often unavailable due to high sequencing costs to obtain comparable coverage. Thus, peak callers which require input controls are impractical for ATAC-seq. Moreover, the direct pile-up of paired-end fragments from ATAC-seq represents both nucleosome-free and nucleosome-bound regions (Fig. 3a). Open chromatin can be detected by piling up short fragments from NFRs or using a shift-extend approach, which tries to count the cutting events smoothed by the extension size (Fig. 3b, right box) [61, 62]. This approach is more generic, as it can be applied to almost all ChIP-seq peak callers and is not affected by the fragment size of data.
Schematic and real ATAC-seq data from core and advanced analyses. a In an ATAC-seq experiment, Tn5 binds and cuts open chromatin and simultaneously ligates adapters. The fragments are sequenced to identify open chromatin regions (black) and footprints (blue). NFR fragments represent the open chromatin, while nucleosome-bound fragments reflect nucleosome positions (gray shaded tracks). b Real ATAC-seq data. Signal tracks are generated from BAM file (Raw) and bias corrected by HINT-ATAC (Bias corrected). Peak sets are generated from three types of peak callers, count-based (red), shape-based (blue), and HMM based (black). For MACS2, two strategies (paired-end and shift-extend) are used. For HMMRATAC, the extended ranges at both sides indicate the nucleosomes. The HINT-ATAC track is footprints detected by HINT-ATAC, while the RUNX1 motif track is the footprints matching RUNX1 motif from JASPAR database. The K562 ChIP-seq track is the RUNX1 ChIP-seq from ENCODE, indicating the footprint detection can recapitulate the real TF binding. The right box illustrates the shift-extend approach. First, it shifts both ends s-bp towards outside, and then extend 2s-bp towards inside. c Illustration of network reconstruction by ATAC-seq data. The presence of TF can be represented by motifs or footprints detected by aforementioned methods. NFR: nucleosome-free region; TF: transcription factor; HMM: hidden Markov model
Popular peak callers for ATAC-seq can be divided into two major categories: count-based or shaped-based. The count-based peak callers employ different statistical methods to compare read distribution shape in a candidate region to a random background. MACS2 [63], HOMER [64], and SICER/epic2 [65,66,67] assume Poisson distribution, while ZINBA [68] assumes zero-inflated negative binomial distribution. F-seq [69] and PeakDEck [70] use kernel density estimation to profile fragment distribution. SPP [71] has no assumption on fragment distribution, but uses a sliding window to calculate scores based on fragment counts from up- and downstream flanking windows. One should keep in mind that some tools, such as F-seq and ZINBA, are not actively maintained and should therefore be used with caution. When applying mixture model clustering to biological replicates, JAMM can determine peak width and boundaries more accurately [72]. In general, count-based methods are easier to interpret and widely used.
Shape-based peak callers are not currently used in ATAC-seq, but they utilize read density profile information directly or indirectly and are believed to improve peak calling in ChIP-seq [73]. PICS [74] models fragment positions other than counts and calculate enrichment score for each candidate region. PolyaPeak [75] ranks peaks using statistics describing peak shape. CLC [76] learns a Gaussian filter for peak shape from positive and negative peaks.
Currently, HMMRATAC is the only peak caller that is exclusive for ATAC-seq [61]. It employs a three-state semi-supervised hidden Markov model (HMM) to simultaneously segment the genome into open chromatin regions with high signal, nucleosomal regions with moderate signals, and background regions with low signals, respectively. Although HMMRATAC is computationally more intensive, it performs better than MACS2 and F-seq and provides additional nucleosome position information at the same time.
Other considerations should include whether the peak caller accounts for Tn5 cleavage bias and how it deals with biological replicates. Similar to DNase-seq, the enzymatic cut by Tn5 will introduce bias due to binding preference [30, 31, 77], which is associated with GC content and should be adjusted when calling peaks [22, 56]. Biological replicates can improve reproducibility and reduce false positive peaks. Most tools can be extended to replicates by either pooling raw reads or combining peak sets from individual samples. Replicates can also be integrated using mixture models [72].
Peak tracks generated by these tools can be visualized in Fig. 3b. Count-based tools behave similarly but are quite different from shape-based tools. Furthermore, the underlying sequence features of these peaks were extracted using neural networks and were shown to recapitulate known TF motifs. This confirmed that TFs play an important role in gene regulation through open, accessible chromatin [78, 79]. Fine tuning of parameters is essential for all aforementioned tools [9, 33], as the width of open chromatin varies [32]. Tools that stitch nearby narrow peaks to form broad peaks such as MACS2, HOMER, and SICER/epic2 are also thought to provide more meaningful results. However, to date, there is no comprehensive benchmark study on peak callers for ATAC-seq, and we suggest using actively supported tools, such as MACS2 and HOMER for peak calling, and if computational resources are sufficient, HMMRATAC could be used for ATAC-seq peak calling.
Advanced analysis
Peaks
Because by its nature ATAC-seq reveals multiple aspects of transcriptional regulation, the third major step involves interpretation at four different levels: peak, motif, nucleosome, and TF footprint. However, only a few tools are designed specifically for ATAC-seq.
Peak differential analysis
Currently, no differential peak analysis tools have been specifically developed for ATAC-seq data analysis. A straightforward approach would be to find the candidate regions (consensus peaks or binned genome), normalize, and count the fragments in these regions and compare with other conditions statistically [80]. This could be achieved manually or using automated tools, such as consensus peak or the sliding window-based tools (Fig. 4b).
Summary of peak calling and peak differential analysis tools. a Peak callers can be divided into count-based, shape-based, and Markov model approaches. They can be further divided by the statistical methods or models used. b Peak differential analysis tools can be divided into peak set-based and sliding window approaches. Peak set-based methods are divided based on the usage of external peak caller and RNA-seq DE packages. Sliding window methods are divided based on statistical methods or models used. ZINB: zero-inflated negative binomial; HMM: hidden Markov model; DE: differential expression; NB: negative binomial
Among the consensus peak-based tools, HOMER, DBChIP [81], and DiffBind [82] rely on RNA-seq differential expression (DE) analysis packages, such as edgeR [83], DESeq [84], or DESeq2 [85]. Thus, they all assume a negative binomial (NB) distribution and require biological replicates to estimate dispersion. It has been suggested to call consensus peaks by pooling all samples to reduce false positive differential peaks which is the default behavior for HOMER [86]; however, DBChIP and DiffBind generate consensus peaks by intersection or union operations. Nevertheless, an intersection operation ignores sample or condition specific peaks, and a union operation often shows lower P values and more false positives [86].
Sliding window approaches do not require pre-generated peak sets. Instead, they evaluate all windows along the binned genome and tend to yield more false positives and require stringent filtering and false discovery rate (FDR) control. PePr [87] and DiffReps [88] use NB test, G-test, or chi-square test, depending on the availability of replicates. For broader peaks, ChIPDiff [89, 90] employs an HMM to account for correlation between adjacent windows. These three tools are independent of RNA-seq DE analysis packages. In contrast, csaw was developed by extending edgeR framework to binned genomes [91]. The sliding window approach is thought to give more unbiased estimates of read count across the genome but requires rigorous FDR control to properly merge adjacent windows.
Currently, most studies assume that ATAC-seq reads in peak regions follow a NB distribution, as is the case for RNA-seq data. However, no shape-based differential analysis tools exist for ATAC-seq data. The peaks contain not only read count information, but also the distribution shape profile. It is especially important for broad peaks, as broad peaks can contain multiple local maxima, and those shifts can indicate biologically relevant perturbations, which could be detected in sliding window or shape-based methods. Although not systematically studied, we believe incorporating shape information will improve differential peak analysis. Nevertheless, considering replicate handling, external peak caller dependency and backend statistical methods, csaw is worth a first try due to its easily explainable edgeR framework.
Peak annotation
After obtaining peak sets, annotation of peaks can associate chromatin accessibility with gene regulation. Normally, peaks are annotated by the nearest genes or regulatory elements. HOMER, ChIPseeker [92], and ChIPpeakAnno [93] are widely used to assign peaks to nearest or overlapping gene, exon, intron, promoter, 5′ untranslated region (UTR), 3′ UTR, and other genomic features. ChIPseeker and ChIPpeakAnno also have abundant visualization functions for interpreting annotation results, such as a pie chart of annotated genomic features (example in Fig. 1d). Typically, peaks from ATAC-seq will represent a mixture of different cis-regulatory elements including enhancers and promoters [12]. After obtaining the list of genomic features such as the nearest genes, functional enrichment analysis can also be performed using databases such as Gene Ontology (GO) [94], KEGG [95], and Reactome [96]. In general, peak annotation generates biological and functionally meaningful results for further investigation.
Motifs
Although peak annotation provides functional interpretation, it does not directly explain the underlying mechanism. Open chromatin can affect transcription through TFs, which facilitate transcription by recognizing and binding to specific sequences on DNA. This sequence is known as a motif and the binding positions are called TF binding sites (TFBS). There are approximately 1600 TFs in human, and more than half have motifs obtained experimentally or computationally [97]. Most TFs require chromatin to be accessible for binding, while certain pioneer TFs can bind to less accessible nucleosomal DNA [98, 99]. TFs regulate transcription through competition with histone or non-histone proteins [100, 101] and cooperation with co-factors [102]. These chromatin accessibility remodeling processes have been reviewed in detail by Klemm, Shipony, and Greenleaf et al. in a recent publication [103]. Thus, understanding motif usage or activity change may help to decipher the underlying regulatory networks, as well as identify key regulators [104]. There are two types of motif or TF-based analysis methods: sequence-based prediction for motif frequency or activity, and footprinting for TF occupancy (discussed in next section).
Motif database and scan
In order to exploit motif information, great efforts have been made to compile databases of motif sequences from either experimental methods or computational predictions. Popular databases such as JASPAR [105] contain multiple species and can be easily retrieved using application programming interfaces (APIs) or Bioconductor packages [106, 107]. To name a few databases, CIS-BP [108] and TRANSFAC [109] contain eukaryotic TF motifs, HOCOMOCO [110] focuses on human and mouse, and RegulonDB [111] is specifically for E. coli. However, there is no central database, which contains comprehensive and consistent motif information, and the discrepancies can arise from differences of original ChIP-seq experiments and the software used to perform de novo motif discovery.
The motif information is mainly stored in text format, for example, as a position weight matrix (PWM). HOMER and Bioconductor packages TFBSTools [112] and motifmatchr [113] are able to search given nucleotide sequences for putative TFBSs using PWM. PWMScan [114] provides a web server for fast motif scanning using a Bowtie indexed genome. Another widely used tool is MEME suite [115, 116], which includes FIMO [117] to search for individual motifs, MAST [118] for aggregating search results from multiple motifs, and MCAST [119] to infer regulatory modules formed by multiple motifs. These tools generate a list of putative TFBSs based on statistical matching. Among them, MEME suite and PWMScan are more accessible owing to their web application interfaces.
Motif enrichment and activity analysis
Based on aforementioned motif search tools, the position and frequency of motifs in each peak region can be obtained and compared to a random background or another condition. HOMER uses the hypergeometric test, while MEME-AME [120] uses the rank-sum test to compare motif frequencies within peaks. MEME-CentriMo [121] further identifies motifs enriched near peak centers. DAStk [62] generates a MD score (motif displacement score) [122]. This is achieved by calculating the ratio of motif occurrence within a small window (150 bp) to a large radius (1500 bp) from each peak center. The MD score can also be compared across different conditions with a Z-test. These methods employ different statistical tests to compare the motif frequency in peaks and the background regions.
Apart from an overrepresentation test, accessibility at each putative TFBS is assumed to associate with TF activity and can be measured by fragment counts. ChromVAR [56] calculates accessibility deviation across multiple conditions for each motif using a Z-score and is adjusted for known technical bias (GC bias, average accessibility and fraction of reads in peaks). It is specifically designed for scATAC-seq data with a large number of cells that could be considered as replicates. However, its performance in bulk ATAC-seq has not been evaluated yet. DiffTF generates a distribution of accessibility fold changes for all TFBSs, adjusted for GC content for each motif and is then compared to a permutated null background to evaluate significance [123, 124]. In summary, MEME-CentriMo is a widely used web application that produces a visual report, while chromVAR can be an alternative in scATAC-seq.
All tools mentioned so far predict putative TFBSs indirectly from sequences found within peak regions. Such TFBSs can contain a significant fraction of false positives and are likely to be incomplete and confounded. This is because not all TFs have identified motifs and TFs from the same family can share very similar motifs [125]. Moreover, the predicted enrichment or activity change could have negligible biological meaning which hampers the interpretation of the sequence-based motif analysis results.
Footprints
Another way to decipher the TF regulation is to use footprints. A footprint in ATAC-seq refers to a pattern where an active TF binds to DNA and prevents Tn5 cleavage within the binding site. This leaves a relative depletion within the open chromatin region (Fig. 3a) [47, 126, 127]. Thus, footprints of actively bound TFs can be used to reconstruct a regulatory network specifically for certain samples.
However, there are hurdles for ATAC-seq footprinting analysis. First, it is important to shift the raw reads in the pre-processing step to account for the 9-bp duplication for accurate footprint detection [9, 33]. Second, due to binding preference of Tn5 [32, 128] and the weak signal of transient TF binding [129], footprint detection is both experimentally and computationally difficult [130]. Great efforts have been made in DNase-seq footprinting, which faces similar challenges except for the difference in enzymatic bias. However, only a few footprinting tools have been tested on ATAC-seq and no systematic evaluation has been performed [48, 131, 132].
Footprinting analysis tools mainly fall into two categories: de novo and motif-centric methods. De novo methods predict all footprint sites across peaks, according to features of a typical footprint pattern (peak-dip-peak). Then these putative footprint sites are used to match known motifs or identify novel motifs. Instead, motif-centric methods require the input of a priori TFBSs and discriminate these sites as bound or unbound using supervised or unsupervised methods (Table 1).
De novo tools
For de novo methods, it is important to mathematically define what is a footprint and denoise the footprint pattern from Tn5 cleavage bias [128, 134]. Boyle et al. [135] proposed an HMM using normalized and smoothed fragment counts at each base to detect different states such as footprint, flanking, and background. HINT, HINT-BC (bias correction for DNase-seq), and recent HINT-ATAC also employ HMM, but only HINT-ATAC corrects for strand-specific Tn5 cleavage bias (Fig. 3b) [130, 133, 134]. An example was shown in Fig. 3b, where footprints detected by HINT-ATAC in a leukemia sample were also validated in a K562 cell line with RUNX1 ChIP-seq. Because these HMM-based methods require supervised training using manually annotated genomic regions, their generalizability in larger datasets needs to be further evaluated. Wellington and Wellington-bootstrap [136, 137] compare the number of Tn5 cuts in flanking and candidate footprint region to find the local minima. Bias correction is not considered by Neph’s method, Boyle’s method, HINT, and Wellington, while DNase2TF and HINT-BC do account for bias correction for DNase-seq [47, 129]. Parameter tuning is a critical consideration and will affect the resultant calls. An optimized pipeline using HINT and Wellington has been described, which evaluates results using area under curve (AUC) analysis considering ChIP-seq binding sites as true positive [48]. In summary, only HINT-ATAC currently handles ATAC-seq-specific bias.
Motif-centric tools
Motif-centric methods focus on a priori TFBSs and consider TF-specific footprint profiles compared to de novo methods. The challenge is to avoid ascertainment bias where TFs with high-quality motifs are enriched.
The unsupervised motif-centric methods classify putative TFBSs as bound or unbound, based on features extracted from genomic regions, e.g., distance to TSS, PWM match score, and sequence conservation score [145, 146], as well as from sequencing reads, e.g., read number and shape distribution around the putative TFBSs [139,140,141, 147]. Among them, CENTIPEDE models read distribution with a multinomial model, and its performance is sensitive to parameters in a TF and cell-type-specific way [133, 139, 143], whereas msCentipede and Romulus account for these heterogeneous footprint profiles [140, 141]. Additionally, msCentipde can model Tn5 bias and Romulus improves performance for low depth data and low-quality motifs. PIQ [147] uses a Gaussian process to model read distribution and can further increase robustness when replicates are provided. The accuracy of unsupervised tools relies heavily on feature selection and construction, thus feature engineering and selection techniques, such as one-hot encoding, binning, and clustering, can be attempted to further improve performance.
In contrast, supervised motif-centric tools require high-quality ChIP-seq to annotate true TFBSs as training data. MILLIPEDE and BinDNase both use logistic regression [142, 143], while DeFCoM uses support vector machine (SVM) and BPAC uses a random forest classifier [131, 144]. Specifically, BinDNase trains a model for each TF separately to account for the TF-specific footprint pattern. The SVM approach used in DeFCoM is more robust to outliers compared to logistic regression [131]. Additionally, DeFCoM was tested on ATAC-seq data and showed slightly decreased performance compared to in DNase-seq with twice the read number. For all supervised tools, performance decreases in cross-TF/cell-type validation, due to variable footprint patterns [142]. This could hamper their application in rare cell populations or heterogeneous cancer samples. An ensemble of larger and more diverse training data was shown to improve footprinting performance [144], and we would also expect ensemble learning to be beneficial, where multiple learners are trained to predict collectively. Moreover, all these tools are trained using DNase-seq data, thus they should be retrained using ATAC-seq data to account for intrinsic bias of different data. In general, modeling TF and cell-type-specific footprint patterns remains difficult due to their substantial variability.
If global TF footprint pattern changes between conditions is of interest, BaGFoot [132] can be employed. It calculates footprint depth and flanking accessibility for all TFs after sequence depth normalization and bias correction. This method is robust to assay type (DNase-seq or ATAC-seq), peak caller, and bias correction methods [132].
Comments on footprinting analysis
There are several caveats for footprinting analysis. First, supervised motif-centric footprinting tools generally outperform unsupervised counterparts and de novo methods, with the trade-off of less generalizability [130, 131]. They have been trained using ChIP-seq and DNase-seq data from specific TFs in specific cell types. Therefore, their context may not be generalizable and applicable to ATAC-seq. Moreover, training data is not always available from the sample of interest, and cross-TF/cell-type prediction should be conducted with caution [131, 144]. Generalizability of these tools to ATAC-seq still requires extensive evaluation. Second, bias correction is important in both DNase-seq and ATAC-seq footprint detection. Recently, the Tn5 preferential motif has been identified and shown to confound some C2H2 zinc finger TFs [128]. Third, there is not a general guideline for minimal ATAC-seq sequencing depth in order to achieve effective footprinting. Although more than 200 million reads per sample are recommended, DeFCoM has been described to work comparably well with fewer sequencing reads [10, 48, 131]. With increasing depth, the improvement of footprinting varies between TFs and cell types due to different binding affinity and turnover [131]. However, saturation analysis is needed to provide reasonable suggestion for experimental design regarding sequencing depth per sample. Fourth, de novo methods still have the advantage for low-quality and novel motifs. Although the evaluation of footprint methods is inconsistent across different studies because of the analysis tools chosen, parameter setting, and evaluation metrics, we believe that HINT-ATAC can be a decent option due to its ATAC-seq-specific bias correction [130, 131]. Additionally, researchers could combine the results from multiple tools to obtain highly reliable footprints. Nevertheless, footprinting analysis in ATAC-seq is useful to understand TF regulation and further reconstruct cell-specific regulatory networks, and therefore requires extensive benchmarking for software comparison and development within specific contexts.
Nucleosome positioning
The nucleosome consists of a histone octamer complexed with approximately 147 bp of DNA (Fig. 3a) and affects TF binding by altering chromatin accessibility [2, 103, 148]. In a standard ATAC-seq library, longer fragments correspond to nucleosome-associated regions (Fig. 3a) [9]. There are analysis tools developed to detect regions where these nucleosomal fragments are enriched. However, Schep et al. showed that nucleosome detection in ATAC-seq was more difficult than in MNase-seq data, due to the deceased read coverage beyond the open chromatin regions [149].
Software developed for MNase-seq, such as DANPOS2, PuFFIN, iNPS, and NucTools, can be applied to ATAC-seq data after filtering for nucleosome-associated fragments [149,150,151,152,153], while NucleoATAC and HMMRATAC are developed specifically for ATAC-seq. NucleoATAC outperformed DANPOS2 by devising a signal score for each base by cross-correlating positional signals with V-plots. A V-plot is a dot-plot to visualize fragment size and midpoint position and is conserved across species [149, 154, 155]. The signal score is normalized and smoothed, and local maxima are found by log-likelihood. HMMRATAC can simultaneously detect open chromatin and nucleosome-associated regions as discussed before (Fig. 3b) [61]. Moreover, DANPOS2 and NucTools can detect the nucleosome occupancy change and position shift between conditions [150, 151]. INPS incorporates a wavelet denoising method while PuFFIN sums up nucleosomal fragment distributions weighted by fragment sizes to identify nucleosomes [152, 153, 156].
However, all these tools suffer from the same underlying drawbacks of a typical ATAC-seq experiment, which is low coverage beyond open chromatin. In the future, new experimental protocols coupled with bioinformatic methods for ATAC-seq will be required to more efficiently and precisely capture nucleosome occupancy. Here, we believe HMMRATAC and NucleoATAC are two useful and specific tools for ATAC-seq nucleosome detection.
Integration with multiomics data to reconstruct regulatory networks
Along with the specified requirements for ATAC-seq data analysis we have articulated so far, the integration of ATAC-seq with other high-throughput sequencing technologies such as RNA-seq and ChIP-seq is gaining increasing interest to understand gene regulation.
Integration with ChIP-seq
Because open chromatin is the pre-requisite for most TFs to bind, ATAC-seq peaks generally overlap with TF ChIP-seq peaks but are often broader. Thus, TF ChIP-seq and ATAC-seq can mutually validate the quality and reliability of each other within the same experimental system [157]. Unique peaks in TF ChIP-seq could indicate pioneer TFs that bind to closed chromatin, which then recruit chromatin remodellers or other TFs and initiate transcription [98, 103]. Analysis based on putative TFBS, such as motif enrichment and footprint detection, can be further improved by incorporating true TF ChIP-seq peaks to reduce false positives [54]. ATAC-seq can also be integrated with histone marker ChIP-seq and is found to positively correlate with active chromatin makers (H3K4me3, H3K4me1, H3K27ac, etc.) and negatively correlate with inactive chromatin markers (H3K27me3) [9, 157, 158]. In conclusion, integrating ChIP-seq and ATAC-seq helps to understand TF and histone facilitated chromatin accessibility changes. We foresee ATAC-seq to be a pioneer assay before specific TF ChIP-seq, due to the ease of the protocol and less sample requirement.
Integration with RNA-seq
Researchers are also interested in qualitatively or quantitatively associating changes in chromatin accessibility with changes in gene expression by RNA-seq. Intuitively, researchers can discover whether DE genes also have significantly differential chromatin accessibility surrounding the respective TSS [159]. Moreover, DE genes can be inferred to be regulated by TF associated with specific motifs or footprints in open chromatin. At the single cell level, Litzenburger et al. attempted to combine scRNA-seq and scATAC-seq to identify the target genes whose expression varies when GATA binding site accessibility changes [160]. Cao et al. used a LASSO regression model to identify distal peaks which account for the target gene expression change [161]. Coupled clustering combining scATAC-seq and scRNA-seq was shown to improve accuracy in subpopulation detection [162]. Integration of ATAC-seq with RNA-seq aids to decipher gene regulation and cellular heterogeneity.
Reconstruction of regulatory networks
While ATAC-seq can simultaneously detect hundreds of TF motif occurrences or footprints, it is possible to reconstruct cell-specific regulatory networks by linking footprints/motifs with downstream genes. Similar approaches have been demonstrated in DNase-seq (Fig. 3c) [104, 163]. However, previous attempts in DNase-seq have been restricted to promoter regions and only investigate TF-TF regulation [104]. Peaks within promoters only constitute a small proportion of all ATAC-seq peaks, while the majority are found in distal enhancers reducing the power to infer regulatory networks [9]. Enhancers can be very distant in a linear genome but spatially proximal (in 3D) to their target genes. This leads to the difficulty of predicting direct target genes of enhancers. Many studies have considered distal peaks as enhancers and linked them to the closest gene akin to a promoter analysis [164,165,166]. With scATAC-seq, Pliner et al. proposed Cicero, which accurately recapitulates co-accessible peaks and links enhancers and promoters to the same target gene. This method has been validated by orthogonal methods [167]. While it has been demonstrated to work in scATAC-seq, it is unclear if this method is applicable to bulk ATAC-seq with much smaller sample sizes. Nevertheless, Cicero is a forerunner in connecting distal enhancers to gene regulation using ATAC-seq.
Although it is possible to reconstruct undirected TF-gene regulatory networks with ATAC-seq alone, the directional regulation can be further inferred as activation or repression when RNA-seq is integrated. Duren et al. proposed a model with paired gene expression and chromatin accessibility (PECA) data to predict the target gene expression as a function of TF expression, chromatin remodeller expression and chromatin accessibility [168]. Miraldi et al. used ATAC-seq-derived binary TF-gene interactions as prior networks, to further refine regulatory networks inferred from RNA-seq data [166]. Berest et al. classified TFs to be activators or repressors based on correlation of TF expression and accessibility at TFBSs across the whole genome [124] with the assumption that accessibility, similar to histone markers, positively correlates with TF expression for activators and negatively for repressors [124, 169]. This method only allows classification in a global manner.
In order to further improve network reconstruction, publicly available ChIP-seq datasets can be integrated to improve the accuracy of footprinting. Incorporating known enhancer-promoter interactions from chromatin conformation data would also be helpful. With the surge of deep learning, it would require more work on feature construction and selection in order to build effective algorithms to predict transcriptional regulatory networks. In summary, integrating ATAC-seq with multiomics data yields biologically meaningful results, which can uncover underlying mechanisms of gene regulation.
Pipelines for ATAC-seq data
There is growing need for integrated pipelines to process ATAC-seq data. Several have been developed but have different focus for downstream analysis by stitching together previously discussed tools.
To name a few, esATAC [170] and CIPHER [171] focus on peak annotation, while GUAVA [172], a graphic user interface (GUI) tool, focuses on differential peak detection as well as functional annotation. ATAC2GRN [48] is another pipeline specifically optimized for footprinting.
These pipelines will provide a helpful and convenient entry for researchers with minimal programming skills to explore ATAC-seq data. However, a general problem for these pipelines is the lack of flexibility for parameter tuning. Most parameters are hard coded empirically because the combination of them increases exponentially with the number of tools, which makes pipelines difficult to modify for any given context. Overall, a pipeline with visualization and user interface will be more appropriate for nonprogrammers to explore the data.
Single-cell ATAC-seq
Enabled by microfluidic, nano-well, and combinatorial indexing technologies, scATAC-seq is now able to measure the chromatin accessibility for thousands of cells with easy protocol at a low cost [33,34,35]. The chromatin accessibility at each base will be binary and the scATAC-seq data will be sparse because in diploid organisms, there are only two copies of DNA. This is a challenge in analyzing scATAC-seq data. Despite the analyses listed for bulk ATAC-seq, another important analysis for single-cell is clustering. A recent benchmarking study from Chen et al. about clustering methods in scATAC-seq showed that SnapATAC, Cusanovich2018 and cisTopic outperformed other methods [23, 173,174,175]. These three methods are featured by workflows combining window-based genome binning, binarization of the accessibility, coverage bias correction, and dimension reduction using principle component analysis, which specifically handle the sparse scATAC-seq data [175]. This study provides a useful insight for future scATAC-seq software development.
New techniques such as scNMT-seq, sci-CAR, and Pi-ATAC were recently developed to measure chromatin accessibility, transcriptome, and proteome simultaneously from exactly the same cell [161, 176, 177]. Data from these experiments could help to deduce the complex interplay between the epigenome, transcriptome, and proteome and help us to understand why different cells behave distinctively. With the advantages of single-cell analysis are clear, there are challenges. Cost and time-efficient single-cell techniques as well as bioinformatic tools remain an area of active research and development.
Future perspectives and concluding remarks
ATAC-seq has developed rapidly over recent years and has become a method of choice to investigate chromatin accessibility. There are now optimized protocols that work with single cells, blood samples, and frozen tissue with improved signal-to-noise ratio [26, 33,34,35, 178]. Despite the progress in protocols, the advancement in bioinformatics analysis tools is slow, with no comprehensive analytical pipeline defined. This imposes a current and ongoing hurdle in the interpretation of ATAC-seq results.
In this review, we have systematically discussed all major steps in an ATAC-seq analysis pipeline for the reader to consider, starting with raw sequencing reads to the endpoint of biological meaningful interpretation. Here, we offer a guide of available tools and suggested steps of analysis to consider to facilitate proper biological interpretation of ATAC-seq data. The alignment and QC steps are similar to RNA-seq and ChIP-seq. As for the peak calling, most ChIP-seq derived tools are compatible with ATAC-seq data. However, a comprehensive benchmarking would help to select appropriate tools and to guide future development of ATAC-seq-specific peak callers. There is growing evidence that improvement or parametrization of current tools can be applied to fit ATAC-seq data.
For downstream interpretation, differential peak analysis can give an overview of the changes of chromatin accessibility. Nevertheless, these changes can arise from both read numbers and the shapes of peaks and can be detected by count-based or sliding window approaches. The performance of these two approaches still requires further evaluation in ATAC-seq and could be specific to particular contexts. In order to infer biological function and related TFs, peak annotation and motif enrichment analysis is a good first pass analysis for initial insight.
Motifs and footprints are direct and indirect indicators of regulatory events respectively. The difficulty in detecting footprints comes from both enzymatic cutting bias and weak signals from transient TFs. Instead of defining footprints with a mathematical formula, recent publications made a good first attempt to embrace the fast development of machine learning algorithms with supervised learning [131, 144]. Moreover, nucleosome detection remains difficult due to an intrinsic weakness of ATAC-seq data where low read coverage beyond peaks is typical. NucleoATAC and HMMRATAC have attempted this; however, large gaps in methods of detection remain in this area.
Another consideration for analysis is on reconstructing gene regulatory networks from ATAC-seq data alone or integrating with multiomics data. This is particularly tempting because ATAC-seq can work with as low as 500 cells and allow the study of well-defined subpopulations especially in developmental biology and clinical samples. ScATAC-seq provides another option to study chromatin biology in heterogeneous cell populations.
In summary, ATAC-seq, an information rich assay, is in great demand for specific bioinformatic analysis tools for further exploitation in analyzing chromatin state, TF footprint, nucleosome position, and regulatory network reconstruction. As a starting point, we suggest researchers can build an effective workflow, by combining FastQC, trimmomatic, and BWA-MEM for pre-analysis, and MACS2 for peak calling. For advanced analysis, we suggest csaw for differential peak analysis, MEME suite for motif detection and enrichment, ChIPseeker for annotation and visualization, HMMRATAC for nucleosome detection, and HINT-ATAC for footprint analysis. If RNA-seq data is available, regulatory networks can be reconstructed using PECA method. However, researchers can always refer to this review for alternative tools for each step and we recommend selecting the tool based on the context of the experimental system and the data collected.
We envisage that this review will encourage researchers to appreciate the complexity and current major hurdles in ATAC-seq data analysis. New ATAC-seq-specific tools and comprehensive benchmarking studies would enable the answering of more biological questions with ATAC-seq in the near future.
Change history
04 May 2023
The correspondence email address has been updated.
References
Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–71.
Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–50.
Human Genome Sequencing Consortium I. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45.
Grewal SIS, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802.
Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605.
Risca VI, Greenleaf WJ. Unraveling the 3D genome: genomics tools for multiscale exploration. Trends Genet. 2015;31:357–72.
Huisinga KL, Brower-Toland B, Elgin SCR. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115:110–22.
Katan-Khaykovich Y, Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005;24:2138–49.
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8.
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;2015:21.29.1–9.
Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010:1–11.
Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82.
Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–22.
Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–85.
Raha D, Hong M, Snyder M. ChIP-seq: a method for global identification of regulatory elements in the genome. Curr Protoc Mol Biol. 2010;91:21.19.1–21.19.14.
Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7.
Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502.
O’Geen H, Echipare L, Farnham PJ. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. Methods Mol Biol. 2011;791:265–86.
Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37.
Cui K, Zhao K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol Biol. 2012;833:413–9.
Schones DE, Cui K, Cuddapah S, Roh T-Y, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98.
Meyer CA, Liu XS. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet. 2014;15:709–21.
Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell. 2018;174:1309–1324.e18.
Fullard JF, Hauberg ME, Bendl J, Egervari G, Cirnaru M-D, Reach SM, et al. An atlas of chromatin accessibility in the adult human brain. Genome Res. 2018;28:1243–52.
Liu C, Wang M, Wei X, Wu L, Xu J, Dai X, et al. An ATAC-seq atlas of chromatin accessibility in mouse tissues. Sci Data. 2019;6:65.
Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–203.
Rendeiro AF, Schmidl C, Strefford JC, Walewska R, Davis Z, Farlik M, et al. Chromatin accessibility maps of chronic lymphocytic leukaemia identify subtype-specific epigenome signatures and transcription regulatory networks. Nat Commun. 2016;7:11938.
Bryois J, Garrett ME, Song L, Safi A, Giusti-Rodriguez P, Johnson GD, et al. Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat Commun. 2018;9:3121.
Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, et al. The chromatin accessibility landscape of primary human cancers. Science. 2018;362:eaav1898.
Reznikoff WS. Transposon Tn 5. Annu Rev Genet. 2008;42:269–86.
Adey A, Morrison HG, Asan XX, Kitzman JO, Turner EH, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119.
Tsompana M, Buck MJ. Chromatin accessibility: a window into the genome. Epigenetics Chromatin. 2014;7:33.
Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–90.
Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 2015;348:910–4.
Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat Commun. 2018;9:3647.
Angarica VE, del Sol A. Bioinformatics tools for genome-wide epigenetic research. Adv Exp Med Biol. 2017;978:489–512.
Chang P, Gohain M, Yen MR, Chen PY. Computational methods for assessing chromatin hierarchy. Comput Struct Biotechnol J. 2018;16:43–53.
Wang Y, Jiang R, Wong WH. Modeling the causal regulatory network by integrating chromatin accessibility and transcriptome data. Natl Sci Rev. 2016;3:240–51.
Andrews S. Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ Accessed 2 Nov 2018.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.j. 2011;17:10.
Schubert M, Lindgreen S, Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9:88.
Jiang H, Lei R, Ding S-W, Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Ma W, Wong WH. The analysis of ChIP-seq data. Methods Enzymol. 2011;497:51–73.
Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90.
Pranzatelli TJF, Michael DG, Chiorini JA. ATAC2GRN: optimized ATAC-seq and DNase1-seq pipelines for rapid and accurate genome regulatory network inference. BMC Genomics. 2018;19:563.
Barozzi I, Bora P, Morelli MJ. Comparative evaluation of DNase-seq footprint identification strategies. Front Genet. 2014;5:278.
Broad Institut. Picard Tools. https://broadinstitute.github.io/picard/ Accessed 2 Nov 2018.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta - Gene Regul Mech. 1819;2012:914–20.
Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep. 2019;9:9354.
Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Ou J, Liu H, Yu J, Kelliher MA, Castilla LH, Lawson ND, et al. ATACseqQC: a bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics. 2018;19:169.
Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. ChromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods. 2017;14:975–8.
Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8.
Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One. 2010;5:e11471.
Thomas R, Thomas S, Holloway AK, Pollard KS. Features that define the best ChIP-seq peak calling algorithms. Brief Bioinform. 2017;18:441–50.
Koohy H, Down TA, Spivakov M, Hubbard T. A comparison of peak callers used for DNase-seq data. PLoS One. 2014;9:e96303.
Tarbell ED, Liu T. HMMRATAC: a Hidden Markov ModeleR for ATAC-seq. Nucleic Acids Res. 2019;47:e91
Tripodi I, Allen M, Dowell R. Detecting differential transcription factor activity from ATAC-seq data. Molecules. 2018;23:1136.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–8.
Xu S, Grullon S, Ge K, Peng W. Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol Biol. 2014;1150:97–111.
Stovner EB, Sætrom P. Epic2 efficiently finds diffuse domains in ChIP-seq data. Bioinformatics. 2019;35:4392-3
Rashid NU, Giresi PG, Ibrahim JG, Sun W, Lieb JD. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol. 2011;12:R67.
Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–8.
McCarthy MT, O’Callaghan CA. PeaKDEck: a kernel density estimator-based peak calling program for DNaseI-seq data. Bioinformatics. 2014;30:1302–4.
Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–9.
Ibrahim MM, Lacadie SA, Ohler U. JAMM: a peak finder for joint analysis of NGS replicates. Bioinformatics. 2015;31:48–55.
Rye MB, Sætrom P, Drabløs F. A manually curated ChIP-seq benchmark demonstrates room for improvement in current peak-finder programs. Nucleic Acids Res. 2011;39:e25.
Zhang X, Robertson G, Krzywinski M, Ning K, Droit A, Jones S, et al. PICS: probabilistic inference for ChIP-seq. Biometrics. 2011;67:151–63.
Wu H, Ji H. PolyaPeak: detecting transcription factor binding sites from ChIP-seq using peak shape information. PLoS One. 2014;9:e89694.
Strino F, Lappe M. Identifying peaks in *-seq data using shape information. BMC Bioinformatics. 2016;17:S206.
Miskimen KLS, Chan R, Haines JL. Assay for transposase-accessible chromatin using sequencing (ATAC-seq) data analysis. Curr Protoc Hum Genet. 2017;92:20.4.1–20.4.13.
Min X, Zeng W, Chen N, Chen T, Jiang R. Chromatin accessibility prediction via convolutional long short-term memory networks with k-mer embedding. Bioinformatics. 2017;33:i92–101.
Liu Q, Xia F, Yin Q, Jiang R. Chromatin accessibility prediction via a hybrid deep convolutional neural network. Bioinformatics. 2018;34:732–8.
Wang J, Zibetti C, Shang P, Sripathi SR, Zhang P, Cano M, et al. ATAC-Seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat Commun. 2018;9:1364.
Liang K, Keleş S. Detecting differential binding of transcription factors with ChIP-seq. Bioinformatics. 2012;28:121–2.
Stark, R and Brown G. DiffBind: differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/devel/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf Accessed 9 Aug 2018.
Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Lun ATL, Smyth GK. De novo detection of differentially bound regions for ChIP-seq data using peaks and windows: controlling error rates correctly. Nucleic Acids Res. 2014;42:e95.
Zhang Y, Lin Y-H, Johnson TD, Rozek LS, Sartor MA. PePr: a peak-calling prioritization pipeline to identify consistent or differential peaks from replicated ChIP-Seq data. Bioinformatics. 2014;30:2568–75.
Shen L, Shao N-Y, Liu X, Maze I, Feng J, Nestler EJ. DiffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8:e65598.
Xu H, Wei C-L, Lin F, Sung W-K. An HMM approach to genome-wide identification of differential histone modification sites from ChIP-seq data. Bioinformatics. 2008;24:2344–9.
Xu H, Sung WK. Identifying differential histone modification sites from ChIP-seq data. Methods Mol Biol. 2012;802:293–303.
Lun ATL, Smyth GK. Csaw: a Bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2016;44:e45.
Yu G, Wang L-G, He Q-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–3.
Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, et al. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237.
The Gene Ontology Consortium. Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–8.
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–80.
Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–55.
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell. 2018;172:650–65.
Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–41.
Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89.
Zabet NR, Adryan B. The effects of transcription factor competition on gene regulation. Front Genet. 2013;4:197.
Joseph SR, Pálfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, et al. Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife. 2017;6:e23326.
Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci. 2008;33:220–9.
Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019;20:207–20.
Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–86.
Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–6.
Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with bioconductor. Nat Methods. 2015;12:115–21.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80.
Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–43.
Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC® and its module TRANSCompel®: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–10.
Kulakovskiy IV, Medvedeva YA, Schaefer U, Kasianov AS, Vorontsov IE, Bajic VB, et al. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013;41:D195–202.
Gama-Castro S, Salgado H, Santos-Zavaleta A, Ledezma-Tejeida D, Muñiz-Rascado L, García-Sotelo JS, et al. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016;44:D133–43.
Tan G, Lenhard B. TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics. 2016;32:1555–6.
Schep Alicia. Fast Motif Matching in R • motifmatchr. https://greenleaflab.github.io/motifmatchr/ Accessed 30 Mar 2019.
Ambrosini G, Groux R, Bucher P. PWMScan: a fast tool for scanning entire genomes with a position-specific weight matrix. Bioinformatics. 2018;34:2483–4.
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8.
Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings Int Conf Intell Syst Mol Biol. 1994;2:28–36.
Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8.
Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54.
Bailey TL, Noble WS. Searching for statistically significant regulatory modules. Bioinformatics. 2003;19:ii16–25.
McLeay RC, Bailey TL. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010;11:165.
Bailey TL, MacHanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012;40:e128.
Azofeifa JG, Allen MA, Hendrix JR, Read T, Rubin JD, Dowell RD. Enhancer RNA profiling predicts transcription factor activity. Genome Res. 2018;28:334–44.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Berest I, Arnold C, Reyes-Palomares A, Palla G, Rasmussen KD, Giles H, et al. Quantification of differential transcription factor activity and multiomics-based classification into activators and repressors: diffTF. Cell Rep. 2019;29:3147–3159.e12.
Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59.
Vierstra J, Stamatoyannopoulos JA. Genomic footprinting. Nat Methods. 2016;13:213–21.
Sung M-H, Baek S, Hager GL. Genome-wide footprinting: ready for prime time? Nat Methods. 2016;13:222–8.
Martins AL, Walavalkar NM, Anderson WD, Zang C, Guertin MJ. Universal correction of enzymatic sequence bias reveals molecular signatures of protein/DNA interactions. Nucleic Acids Res. 2018;46:e9.
Sung M-H, Guertin MJ, Baek S, Hager GL. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell. 2014;56:275–85.
Gusmao EG, Allhoff M, Zenke M, Costa IG. Analysis of computational footprinting methods for DNase sequencing experiments. Nat Methods. 2016;13:303–9.
Quach B, Furey TS. DeFCoM: analysis and modeling of transcription factor binding sites using a motif-centric genomic footprinter. Bioinformatics. 2017;33:956–63.
Baek S, Goldstein I, Hager GL. Bivariate genomic footprinting detects changes in transcription factor activity. Cell Rep. 2017;19:1710–22.
Gusmao EG, Dieterich C, Zenke M, Costa IG. Detection of active transcription factor binding sites with the combination of DNase hypersensitivity and histone modifications. Bioinformatics. 2014;30:3143–51.
Li Z, Schulz MH, Look T, Begemann M, Zenke M, Costa IG. Identification of transcription factor binding sites using ATAC-seq. Genome Biol. 2019;20:45.
Boyle AP, Song L, Lee B-K, London D, Keefe D, Birney E, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–64.
Piper J, Elze MC, Cauchy P, Cockerill PN, Bonifer C, Ott S. Wellington: a novel method for the accurate identification of digital genomic footprints from DNase-seq data. Nucleic Acids Res. 2013;41:e201.
Piper J, Assi SA, Cauchy P, Ladroue C, Cockerill PN, Bonifer C, et al. Wellington-bootstrap: differential DNase-seq footprinting identifies cell-type determining transcription factors. BMC Genomics. 2015;16:1000.
Hosoya T, D’Oliveira Albanus R, Hensley J, Myers G, Kyono Y, Kitzman J, et al. Global dynamics of stage-specific transcription factor binding during thymocyte development. Sci Rep. 2018;8:5605.
Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–55.
Raj A, Shim H, Gilad Y, Pritchard JK, Stephens M. MsCentipede: modeling heterogeneity across genomic sites and replicates improves accuracy in the inference of transcription factor binding. PLoS One. 2015;10:e0138030.
Jankowski A, Tiuryn J, Prabhakar S. Romulus: robust multi-state identification of transcription factor binding sites from DNase-seq data. Bioinformatics. 2016;32:2419–26.
Kähärä J, Lähdesmäki H. BinDNase: a discriminatory approach for transcription factor binding prediction using DNase I hypersensitivity data. Bioinformatics. 2015;31:2852–9.
Luo K, Hartemink AJ. Using DNase digestion data to accurately identify transcription factor binding sites. Pac Symp Biocomput. 2013:80–91.
Liu S, Zibetti C, Wan J, Wang G, Blackshaw S, Qian J. Assessing the model transferability for prediction of transcription factor binding sites based on chromatin accessibility. BMC Bioinformatics. 2017;18:355.
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21.
Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50.
Sherwood RI, Hashimoto T, O’Donnell CW, Lewis S, Barkal AA, van Hoff JP, et al. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat Biotechnol. 2014;32:171–8.
Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–5.
Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, Greenleaf WJ. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 2015;25:1757–70.
Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, et al. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res. 2013;23:341–51.
Vainshtein Y, Rippe K, Teif VB. NucTools: analysis of chromatin feature occupancy profiles from high-throughput sequencing data. BMC Genomics. 2017;18:158.
Chen W, Liu Y, Zhu S, Green CD, Wei G, Han J-DJ. Improved nucleosome-positioning algorithm iNPS for accurate nucleosome positioning from sequencing data. Nat Commun. 2014;5:4909.
Polishko A, Bunnik EM, Le Roch KG, Lonardi S. PuFFIN - a parameter-free method to build nucleosome maps from paired-end reads. BMC Bioinformatics. 2014;15:S11.
Henikoff JG, Belsky JA, Krassovsky K, MacAlpine DM, Henikoff S. Epigenome characterization at single base-pair resolution. Proc Natl Acad Sci U S A. 2011;108:18318–23.
Zentner GE, Henikoff S. Surveying the epigenomic landscape, one base at a time. Genome Biol. 2012;13:250.
Zhang Y, Shin H, Song JS, Lei Y, Liu XS. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-seq. BMC Genomics. 2008;9:537.
Jiang S, Mortazavi A. Integrating ChIP-seq with other functional genomics data. Brief Funct Genomics. 2018;17:104–15.
Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705.
Ackermann AM, Wang Z, Schug J, Naji A, Kaestner KH. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol Metab. 2016;5:233–44.
Litzenburger UM, Buenrostro JD, Wu B, Shen Y, Sheffield NC, Kathiria A, et al. Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol. 2017;18:15.
Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ, et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361:1380–5.
Duren Z, Chen X, Zamanighomi M, Zeng W, Satpathy AT, Chang HY, et al. Integrative analysis of single-cell genomics data by coupled nonnegative matrix factorizations. Proc Natl Acad Sci U S A. 2018;115:7723–8.
Sullivan AM, Bubb KL, Sandstrom R, Stamatoyannopoulos JA, Queitsch C. DNase I hypersensitivity mapping, genomic footprinting, and transcription factor networks in plants. Curr Plant Biol. 2015;3–4:40–7.
Karwacz K, Miraldi ER, Pokrovskii M, Madi A, Yosef N, Wortman I, et al. Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat Immunol. 2017;18:412–21.
Ramirez RN, El-Ali NC, Mager MA, Wyman D, Conesa A, Mortazavi A. Dynamic gene regulatory networks of human myeloid differentiation. Cell Syst. 2017;4:416–429.e3.
Miraldi ER, Pokrovskii M, Watters A, Castro DM, De Veaux N, Hall JA, et al. Leveraging chromatin accessibility for transcriptional regulatory network inference in T helper 17 cells. Genome Res. 2019;29:449–63.
Pliner HA, Packer JS, McFaline-Figueroa JL, Cusanovich DA, Daza RM, Aghamirzaie D, et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol Cell. 2018;71:858–871.e8.
Duren Z, Chen X, Jiang R, Wang Y, Wong WH. Modeling gene regulation from paired expression and chromatin accessibility data. Proc Natl Acad Sci. 2017;114:E4914–23.
Grubert F, Zaugg JB, Kasowski M, Ursu O, Spacek DV, Martin AR, et al. Genetic control of chromatin states in humans involves local and distal chromosomal interactions. Cell. 2015;162:1051–65.
Wei Z, Zhang W, Fang H, Li Y, Wang X. EsATAC: an easy-to-use systematic pipeline for ATAC-seq data analysis. Bioinformatics. 2018;34:2664–5.
Guzman C, D’Orso I. CIPHER: a flexible and extensive workflow platform for integrative next-generation sequencing data analysis and genomic regulatory element prediction. BMC Bioinformatics. 2017;18:363.
Divate M, Cheung E. GUAVA: a graphical user Interface for the analysis and visualization of ATAC-seq data. Front Genet. 2018;9:250.
Bravo González-Blas C, Minnoye L, Papasokrati D, Aibar S, Hulselmans G, Christiaens V, et al. CisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat Methods. 2019;16:397–400.
Fang R, Preissl S, Hou X, Lucero J, Wang X, Motamedi A, et al. Fast and accurate clustering of single cell epigenomes reveals Cis-regulatory elements in rare cell types. bioRxiv. 2019.
Chen H, Lareau C, Andreani T, Vinyard ME, Garcia SP, Clement K, et al. Assessment of computational methods for the analysis of single-cell ATAC-seq data. Genome Biol. 2019;20:241.
Clark SJ, Argelaguet R, Kapourani C-A, Stubbs TM, Lee HJ, Alda-Catalinas C, et al. ScNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun. 2018;9:781.
Chen X, Litzenburger UM, Wei Y, Schep AN, LaGory EL, Choudhry H, et al. Joint single-cell DNA accessibility and protein epitope profiling reveals environmental regulation of epigenomic heterogeneity. Nat Commun. 2018;9:4590.
Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–62.
Acknowledgements
We are grateful to Dr. Christina Makhlouf, Mr. Graham Magor, and Dr. Le Ying for critical discussion and feedback, and Mr. Zihao Deng for the figure design.
Peer review information
Yixin Yao was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 1.
Funding
FY acknowledges funding support from Monash University (Monash Graduate Scholarship and Monash International Postgraduate Research Scholarship).
Availability of data and materials
The raw sequencing data to produce the fragment size distribution, TSS enrichment plot, and peak tracks from Corces et al. [26] is available in SRA under accession SRR2920587.
Author information
Authors and Affiliations
Contributions
FY proposed the project and drafted the manuscript. NW revised the manuscript. NW, DP, and DC supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Review history. (DOCX 21 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yan, F., Powell, D.R., Curtis, D.J. et al. From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis. Genome Biol 21, 22 (2020). https://doi.org/10.1186/s13059-020-1929-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13059-020-1929-3