-
PDF
- Split View
-
Views
-
Cite
Cite
Douglas G. Howe, Yvonne M. Bradford, Tom Conlin, Anne E. Eagle, David Fashena, Ken Frazer, Jonathan Knight, Prita Mani, Ryan Martin, Sierra A. Taylor Moxon, Holly Paddock, Christian Pich, Sridhar Ramachandran, Barbara J. Ruef, Leyla Ruzicka, Kevin Schaper, Xiang Shao, Amy Singer, Brock Sprunger, Ceri E. Van Slyke, Monte Westerfield, ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D854–D860, https://doi.org/10.1093/nar/gks938
Close - Share Icon Share
Abstract
ZFIN, the Zebrafish Model Organism Database (http://zfin.org), is the central resource for zebrafish genetic, genomic, phenotypic and developmental data. ZFIN curators manually curate and integrate comprehensive data involving zebrafish genes, mutants, transgenics, phenotypes, genotypes, gene expressions, morpholinos, antibodies, anatomical structures and publications. Integrated views of these data, as well as data gathered through collaborations and data exchanges, are provided through a wide selection of web-based search forms. Among the vertebrate model organisms, zebrafish are uniquely well suited for rapid and targeted generation of mutant lines. The recent rapid production of mutants and transgenic zebrafish is making management of data associated with these resources particularly important to the research community. Here, we describe recent enhancements to ZFIN aimed at improving our support for mutant and transgenic lines, including (i) enhanced mutant/transgenic search functionality; (ii) more expressive phenotype curation methods; (iii) new downloads files and archival data access; (iv) incorporation of new data loads from laboratories undertaking large-scale generation of mutant or transgenic lines and (v) new GBrowse tracks for transgenic insertions, genes with antibodies and morpholinos.
INTRODUCTION
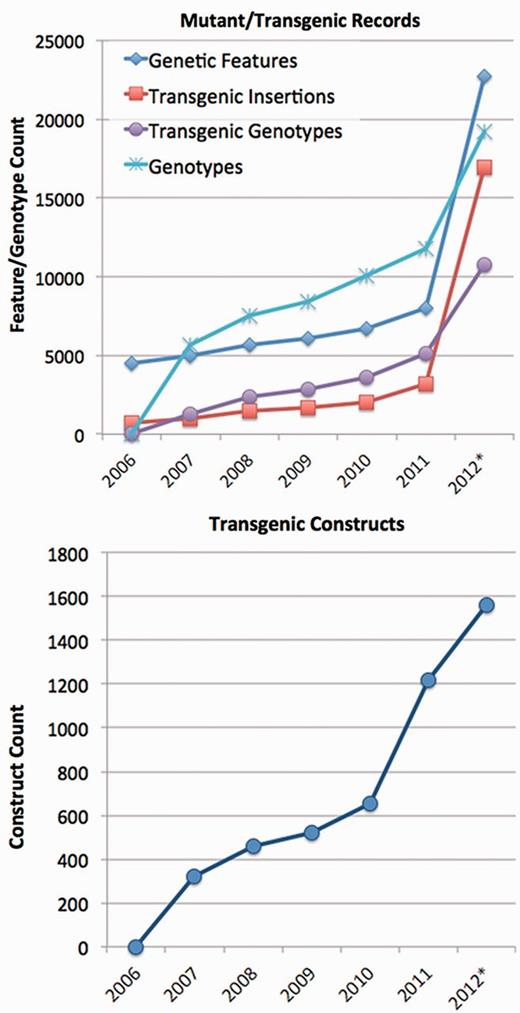
ZFIN (http://zfin.org) is a curated database of zebrafish genetic and genomic data including genes, mutants, transgenic lines and constructs, genotypes, phenotypes, gene expression, anatomical structures, orthology, nucleotide and protein sequences and reagents such as morpholinos and antibodies. Table 1 summarizes ZFIN data contents as of August 2012. A table illustrating growth of these data at ZFIN since 1998 can be accessed on the ZFIN web site at http://zfin.org/zf_info/zfin_stats.html. It is apparent from these data that mutant and transgenic lines and transgenic constructs are the most rapidly growing data types in ZFIN (Figure 1). This is not surprising. As technology has evolved, it has become possible for laboratories to generate large numbers of mutant and transgenic lines at lower cost. To maximize utilization of these resources by the zebrafish research community, it is increasingly important for ZFIN to load, store and retrieve information about large numbers of mutants and transgenics. To meet this need, ZFIN has increased support for storing, searching and displaying information about transgenic constructs and lines, and we have increased the expressivity of our phenotype curation syntax.
In the past 2 years, mutant and transgenic features (Genetic Features) as well as transgenic constructs have been among the most rapidly growing data types at ZFIN. *2012 data through 8 August 2012.
Summary of data content at ZFIN as of 8 August 2012
| Data Type . | 2012a . |
|---|---|
| Genes | |
| Gene records | 32 893 |
| Genes on assembly | 19 332 |
| Transcripts | 31 913 |
| ESTs/cDNAs | 34 865 |
| Full-length cDNA clones (ZGC) | 17 191 |
| Genetics | |
| Genetic features | 22 738 |
| Transgenic insertions | 16 922 |
| Transgenic constructs | 1562 |
| Transgenic genotypes | 10 787 |
| Genotypes | 19 237 |
| Functional annotation | |
| Genes with any GO annotation | 18 051 |
| Genes with IEA GO annotation | 14 569 |
| Genes with non-IEA GO | 8898 |
| Total GO annotations | 139 639 |
| Reagents | |
| Morpholinos | 5035 |
| Antibodies | 916 |
| Expression and phenotypes | |
| Gene expression patterns | 59 829 |
| Images | 83 538 |
| Anatomical structures | 2760 |
| Genomics | |
| Mapped markers | 38 434 |
| Links to other databases | 648 395 |
| Community information | |
| Publications | 16 671 |
| Researchers | 6130 |
| Laboratories | 782 |
| Companies | 148 |
| Orthology | |
| Genes with curated human orthology | 12 601 |
| Genes with curated mouse orthology | 9724 |
| Data Type . | 2012a . |
|---|---|
| Genes | |
| Gene records | 32 893 |
| Genes on assembly | 19 332 |
| Transcripts | 31 913 |
| ESTs/cDNAs | 34 865 |
| Full-length cDNA clones (ZGC) | 17 191 |
| Genetics | |
| Genetic features | 22 738 |
| Transgenic insertions | 16 922 |
| Transgenic constructs | 1562 |
| Transgenic genotypes | 10 787 |
| Genotypes | 19 237 |
| Functional annotation | |
| Genes with any GO annotation | 18 051 |
| Genes with IEA GO annotation | 14 569 |
| Genes with non-IEA GO | 8898 |
| Total GO annotations | 139 639 |
| Reagents | |
| Morpholinos | 5035 |
| Antibodies | 916 |
| Expression and phenotypes | |
| Gene expression patterns | 59 829 |
| Images | 83 538 |
| Anatomical structures | 2760 |
| Genomics | |
| Mapped markers | 38 434 |
| Links to other databases | 648 395 |
| Community information | |
| Publications | 16 671 |
| Researchers | 6130 |
| Laboratories | 782 |
| Companies | 148 |
| Orthology | |
| Genes with curated human orthology | 12 601 |
| Genes with curated mouse orthology | 9724 |
A full table showing growth of these data on an annual basis since 1998 can be found by using the ‘Statistics’ link on the ZFIN home page. aData through 8 August 2012. EST: Expressed sequence Tag; IEA: Inferred from Electronic Annotation.
Summary of data content at ZFIN as of 8 August 2012
| Data Type . | 2012a . |
|---|---|
| Genes | |
| Gene records | 32 893 |
| Genes on assembly | 19 332 |
| Transcripts | 31 913 |
| ESTs/cDNAs | 34 865 |
| Full-length cDNA clones (ZGC) | 17 191 |
| Genetics | |
| Genetic features | 22 738 |
| Transgenic insertions | 16 922 |
| Transgenic constructs | 1562 |
| Transgenic genotypes | 10 787 |
| Genotypes | 19 237 |
| Functional annotation | |
| Genes with any GO annotation | 18 051 |
| Genes with IEA GO annotation | 14 569 |
| Genes with non-IEA GO | 8898 |
| Total GO annotations | 139 639 |
| Reagents | |
| Morpholinos | 5035 |
| Antibodies | 916 |
| Expression and phenotypes | |
| Gene expression patterns | 59 829 |
| Images | 83 538 |
| Anatomical structures | 2760 |
| Genomics | |
| Mapped markers | 38 434 |
| Links to other databases | 648 395 |
| Community information | |
| Publications | 16 671 |
| Researchers | 6130 |
| Laboratories | 782 |
| Companies | 148 |
| Orthology | |
| Genes with curated human orthology | 12 601 |
| Genes with curated mouse orthology | 9724 |
| Data Type . | 2012a . |
|---|---|
| Genes | |
| Gene records | 32 893 |
| Genes on assembly | 19 332 |
| Transcripts | 31 913 |
| ESTs/cDNAs | 34 865 |
| Full-length cDNA clones (ZGC) | 17 191 |
| Genetics | |
| Genetic features | 22 738 |
| Transgenic insertions | 16 922 |
| Transgenic constructs | 1562 |
| Transgenic genotypes | 10 787 |
| Genotypes | 19 237 |
| Functional annotation | |
| Genes with any GO annotation | 18 051 |
| Genes with IEA GO annotation | 14 569 |
| Genes with non-IEA GO | 8898 |
| Total GO annotations | 139 639 |
| Reagents | |
| Morpholinos | 5035 |
| Antibodies | 916 |
| Expression and phenotypes | |
| Gene expression patterns | 59 829 |
| Images | 83 538 |
| Anatomical structures | 2760 |
| Genomics | |
| Mapped markers | 38 434 |
| Links to other databases | 648 395 |
| Community information | |
| Publications | 16 671 |
| Researchers | 6130 |
| Laboratories | 782 |
| Companies | 148 |
| Orthology | |
| Genes with curated human orthology | 12 601 |
| Genes with curated mouse orthology | 9724 |
A full table showing growth of these data on an annual basis since 1998 can be found by using the ‘Statistics’ link on the ZFIN home page. aData through 8 August 2012. EST: Expressed sequence Tag; IEA: Inferred from Electronic Annotation.
MUTANT AND TRANSGENIC DATA
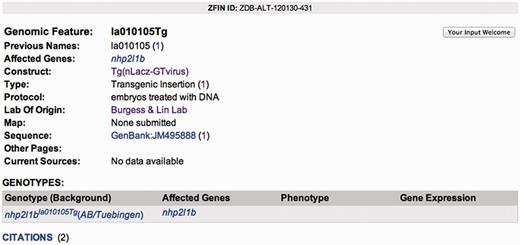
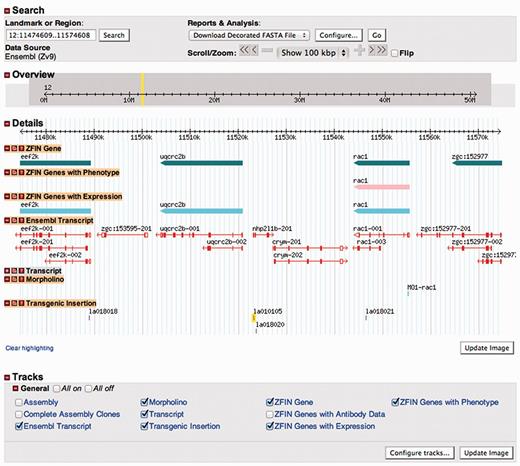
Zebrafish have become one of the pre-eminent model organisms for studies of gene function aimed at understanding human development and disease. One reason for this is the growing ease with which large numbers of zebrafish mutants and transgenics can be created and analyzed. ZFIN has recently developed collaborations with the laboratories of Shawn Burgess and Shuo Lin, Steve Ekker and Derek Stemple (The Zebrafish Mutation Project, ZMP, hosted at the Sanger Institute) to load data from their large-scale mutant/transgenic fish resources (1–4). To make these resources widely accessible to the zebrafish research community, thier data has been annotated and loaded into the ZFIN database, while the fish are made available to the research community by the Zebrafish International Resource Center. Each mutant or transgenic integration site is given an official genomic feature designation (allele number) and is annotated with the affected gene, the construct utilized, the type of mutation, the protocol used, the laboratory of origin, mapping information, associated GenBank accession numbers, current sources and genotypes in which the genomic feature can be found. For transgenic fish, the construct utilized is curated to include details about the promoters, expressed genes and distinctive features such as loxP sites, etc. Genomic features can be searched from the Mutants/Morphants/Transgenics search form and the annotated data can be accessed on the Genomic Feature page (Figure 2). In addition, transgenic insertions for which we have integration site sequence information have a dedicated track in the ZFIN genome browser, GBrowse (Figure 3). The GBrowse view for transgenic insertions is important because it provides a simple graphical view of where a transgenic insertion is located within a gene’s structure. The closer an insertion is to the start site of a gene, the more likely it is to produce a mutant phenotype, information that can be critical in helping researchers select transgenic insertions of interest. To date, ZFIN has loaded 13 192 viral insertions and 4435 genotypes from the Burgess and Lin project, 63 genomic features and 96 genotypes from the Ekker laboratory project, and 881 genomic features and 881 genotypes from the two most recent ZMP data submissions (Table 3). In 2010, the Sanger Institute provided ZFIN with phenotype and image data for 102 of 249 mutants that had been added to ZFIN in earlier loads (5–8). The Sanger Institute will continue to provide ZFIN with phenotype and image data from the ongoing ZMP project.
The Genomic Feature page provides detailed information about the genomic feature. This is an example of a viral insertion feature including affected gene, previous name, associated construct, type of mutation, protocol used, laboratory of origin, mapping information, associated GenBank accessions, current sources and genotypes.
Transgenic insertions now have a dedicated GBrowse track. Like other items shown in Gbrowse, transgenic insertions shown on this track are linked to their feature records in ZFIN.
Ontologies used to describe zebrafish phenotype
| Ontologya . | ID prefix . | Example terms . |
|---|---|---|
| Phenotypic quality | PATO | Yellow, dorsalized, increased rate, fused with, absent |
| Zebrafish anatomy and development | ZFA, ZFS | Posterior lateral line, Rohon-Beard neuron, pancreas, segmentation: 10–13 somites, larval: Day 4 |
| Gene Ontology | GO | Brain morphogenesis, transferase activity, cilium |
| Mouse pathology (neoplasm branch) | MPATH | Carcinoma, hepatoblastoma, spermatocytic seminoma |
| Spatial ontology | BSPO | Anterior/posterior axis, left side, dorsal region |
| Relation ontology | RO | Is_a, part_of, develops_from |
| Ontologya . | ID prefix . | Example terms . |
|---|---|---|
| Phenotypic quality | PATO | Yellow, dorsalized, increased rate, fused with, absent |
| Zebrafish anatomy and development | ZFA, ZFS | Posterior lateral line, Rohon-Beard neuron, pancreas, segmentation: 10–13 somites, larval: Day 4 |
| Gene Ontology | GO | Brain morphogenesis, transferase activity, cilium |
| Mouse pathology (neoplasm branch) | MPATH | Carcinoma, hepatoblastoma, spermatocytic seminoma |
| Spatial ontology | BSPO | Anterior/posterior axis, left side, dorsal region |
| Relation ontology | RO | Is_a, part_of, develops_from |
aOntology files can be downloaded from The Open Biological and Biomedical Ontologies Foundry; http://obofoundry.org.
Ontologies used to describe zebrafish phenotype
| Ontologya . | ID prefix . | Example terms . |
|---|---|---|
| Phenotypic quality | PATO | Yellow, dorsalized, increased rate, fused with, absent |
| Zebrafish anatomy and development | ZFA, ZFS | Posterior lateral line, Rohon-Beard neuron, pancreas, segmentation: 10–13 somites, larval: Day 4 |
| Gene Ontology | GO | Brain morphogenesis, transferase activity, cilium |
| Mouse pathology (neoplasm branch) | MPATH | Carcinoma, hepatoblastoma, spermatocytic seminoma |
| Spatial ontology | BSPO | Anterior/posterior axis, left side, dorsal region |
| Relation ontology | RO | Is_a, part_of, develops_from |
| Ontologya . | ID prefix . | Example terms . |
|---|---|---|
| Phenotypic quality | PATO | Yellow, dorsalized, increased rate, fused with, absent |
| Zebrafish anatomy and development | ZFA, ZFS | Posterior lateral line, Rohon-Beard neuron, pancreas, segmentation: 10–13 somites, larval: Day 4 |
| Gene Ontology | GO | Brain morphogenesis, transferase activity, cilium |
| Mouse pathology (neoplasm branch) | MPATH | Carcinoma, hepatoblastoma, spermatocytic seminoma |
| Spatial ontology | BSPO | Anterior/posterior axis, left side, dorsal region |
| Relation ontology | RO | Is_a, part_of, develops_from |
aOntology files can be downloaded from The Open Biological and Biomedical Ontologies Foundry; http://obofoundry.org.
Basic search functionality for transgenic constructs is available using the ZFIN Genes/Markers/Clones search. ZFIN currently contains records for >1500 distinct transgenic constructs. In the near future, we plan to provide increased support for transgenic constructs, including construct images and a dedicated transgenic construct search that supports queries for specific promoters, expressed genes and distinctive features such as loxP sites and reporter expression patterns.
MUTANT AND TRANSGENIC SEARCH
Recognizing the importance of searches for the rapidly increasing volume of mutant, morphant and transgenic fish, we released a new version of Mutant/Morphant/Transgenic search in May 2012. Changes to the Mutant/Morphant/Transgenic search interface include the ability to search using more than one term, the ability to use Gene Ontology (GO) terms for phenotype, and improved search filters. In addition, the speed of the search has improved significantly by using a backend data mart.
Searching for multiple terms is a web standard, and we are working hard to bring this ability to ZFIN users. The first step in this process was to provide a search for multiple genes and/or alleles at the same time. This is critical when searching for double mutants, a mutant injected with a morpholino, or a transgenic fish expressing GFP under the control of regulatory sequences from specific genes (e.g. pax2a). Lucene text indexes are now embedded in our database to support multi-term searching and to increase the speed of the search. Transforming curated data into a data mart utilizing Lucene text indexes de-normalizes the data so that sorting happens on demand, result lists can be broken down by feature and affected gene without extra queries and figure counts can be displayed without real-time computation.
Although ZFIN has supported searches for mutants with phenotypes affecting specific anatomical structures (AO) for some time, GO biological process (GOBP) terms used in phenotype annotations were not searchable until recently. GOBP terms are an integral part of curated mutant phenotypes at ZFIN, and it is now possible to choose from both GOBP and AO terms in the auto-complete phenotype search box when searching for mutant phenotypes. Additionally, we updated the search filters to allow results to include exclusively or exclude specifically fish that use morpholinos or are transgenic.
Noticeable improvements to the search results include faster results return and improved default ordering of results, column sorting options, a display of constructs for transgenic fish and a link to gene expression data for each fish. Results are returned with what ZFIN considers to be the ‘best match’ returned first. ‘Best match’ is calculated by looking first for exact text matches of the search string, and then ordering results based on a biologically defined set of criteria including number of mutations in a particular fish and the mutation type. For example, simple mutants (ones with single point mutations) are returned with greater priority than mutants with multiple genes affected.
As ZFIN’s ‘best match’ algorithm was improved, results can now be ordered in various ways. If results are more interesting when sorted by affected gene, then results can be sorted alphabetically by mutated/morpholino-affected gene. Likewise, if results are more useful when ordered by allele designation or morpholino, results can be sorted by line designation/morpholino name.
Finding information for specific transgenic fish is also easier now with the addition of the construct column. Not only is it obvious which constructs were used in each fish but also information about the constructs is one click away on the construct page.
Finally, gene expression data for the fish are available directly from the search results. To determine which genes have been recorded as expressed, or not expressed in mutant, morphant or transgenic fish, click on the summarized ‘figures’ link for each fish.
REPRESENTATION OF PHENOTYPES
ZFIN curators manually curate phenotype data for genotypes containing mutant alleles and/or morpholinos. In addition to capturing data from publications, collaborations with research laboratories result in direct submission of phenotype data to the database. We use biological ontologies (controlled vocabularies) to construct phenotype statements (Table 2). This method improves consistency, facilitates searches and allows computation across disparate data sets both within ZFIN as well as across model organisms. At ZFIN, phenotypes are represented using the EQ syntax to create phenotype statements, where E represents the entity (an anatomical structure or GOBP) and Q is the phenotypic quality from the Phenotypic Quality ontology, PATO (e.g. increased size; PATO:0000586, decreased rate; PATO:0000911) (9). In addition, a phenotype statement also includes an abnormal/normal tag or modifier. The ‘normal or recovered’ tag is used when the annotation of a normal phenotype is notable or when the annotation represents a recovered normal phenotype, such as that resulting from the addition of a morpholino to a mutant or the creation of a complex mutant genotype. When a more detailed entity description is needed and a representative term is not available from one of the ontologies, ontology terms can be combined to post-compose a more specific term. For instance, the EQ statement ‘trabecula communis chondrocyte disorganized, abnormal’ combines two anatomy terms, trabecula communis (superterm) and chondrocyte (subterm), to create a more specific entity representing a chondrocyte that is part of the trabecula communis. Phenotype statements are displayed to enhance user readability but have underlying relationships that define the syntax of the annotations. For instance, an EQ statement uses the relationship inheres_in and translates as ‘the quality type, which inheres_in the entity.’ For post-composition, an inferred part_of relationship exists between the subterm and superterm.
Number of genomic features and genotypes loaded into ZFIN in ongoing collaborations. ENU: N-ethyl-N-nitrosourea
| Laboratory . | Number of genomic features in ZFIN . | Number of genotypes in ZFIN . |
|---|---|---|
| S. Burgess and S. Lin | 13 192 viral insertions | 4435 |
| S. Ekker | 63 gene breaking transposon insertions | 96 |
| Zebrafish mutation resource | 881 ENU-induced point mutations | 881 |
| Laboratory . | Number of genomic features in ZFIN . | Number of genotypes in ZFIN . |
|---|---|---|
| S. Burgess and S. Lin | 13 192 viral insertions | 4435 |
| S. Ekker | 63 gene breaking transposon insertions | 96 |
| Zebrafish mutation resource | 881 ENU-induced point mutations | 881 |
Number of genomic features and genotypes loaded into ZFIN in ongoing collaborations. ENU: N-ethyl-N-nitrosourea
| Laboratory . | Number of genomic features in ZFIN . | Number of genotypes in ZFIN . |
|---|---|---|
| S. Burgess and S. Lin | 13 192 viral insertions | 4435 |
| S. Ekker | 63 gene breaking transposon insertions | 96 |
| Zebrafish mutation resource | 881 ENU-induced point mutations | 881 |
| Laboratory . | Number of genomic features in ZFIN . | Number of genotypes in ZFIN . |
|---|---|---|
| S. Burgess and S. Lin | 13 192 viral insertions | 4435 |
| S. Ekker | 63 gene breaking transposon insertions | 96 |
| Zebrafish mutation resource | 881 ENU-induced point mutations | 881 |
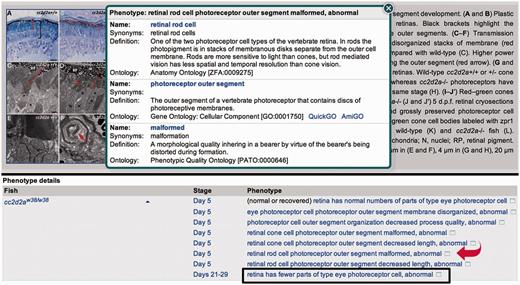
We have introduced several enhancements to zebrafish phenotype annotation during the past year. To increase the detail of phenotype annotations, we now use two additional ontologies for post-composition with zebrafish anatomy terms: the Spatial Ontology and the neoplasm branch of the Mouse Pathology Ontology (MPATH) (10). The Spatial Ontology supports representation of phenotypes for refined sub-regions of anatomical structures (dorsal spinal cord for example), and MPATH neoplasm terms are used to describe cancer phenotypes. Another new addition to ZFIN is the ability to compose and display phenotype statements using relational quality terms from PATO. A relational quality is a type of quality that requires an additional entity, i.e. those expressed in an EQE statement. An example would be the relational term ‘fused with’ used to describe cyclopia with the EQE statement ‘eye fused with eye, abnormal’. With post-composed entities and the use of a relational quality, a complex phenotype statement can be constructed that expresses a high degree of detail, e.g. ‘midbrain posterior region has fewer parts of type dopaminergic neuron axon’. When viewing a phenotype statement at ZFIN, you can find more information about the component ontology terms by clicking the icon at the end of the statement that opens a pop-up window (Figure 4). You can click again to close the pop-up, or you can click a hyperlinked term name to go to a term detail page where you can explore the ontology. The zebrafish anatomy term detail pages include expression and phenotype data associated with the structure, as well as images when available. Links to phenotype data are located on a number of ZFIN pages, including Gene, Genomic Feature and Mutant/Morphant/Tg search result pages; complete phenotype statements are displayed on Figure, Genotype, Anatomy Details, Phenotype Summary and Phenotype Statement pages. Access to the full set of phenotype data is available for download from the ZFIN Data Reports page (http://zfin.org/downloads), which has recently been updated.
Example of a figure page at ZFIN with phenotype summary statements. Clicking the complete hyperlinked phenotype statement opens a Phenotype Statement summary page (not shown). Clicking the icon at the end of the statement (red arrow) opens a pop-up window with additional ontology term information as shown. Click on a hyperlinked term name from the pop-up to redirect to the term detail page for additional information and the ability to navigate the ontology. Represented are examples of a ‘normal or recovered’ phenotype statement, the use of a relational quality (boxed phenotype statement) and post-composition, which is displayed in the pop-up where the AO term retinal rod cell is post-composed with the GO cellular component term photoreceptor outer segment.
DATA DOWNLOADS
In addition to those who access data at ZFIN through the zfin.org web site, a growing segment of the research community needs direct access to downloads of full data sets with which they can perform more comprehensive bioinformatic analyses. The majority of data in the ZFIN database are freely available from the downloads page (https://zfin.org/downloads) that is accessed from a link on the ZFIN home page. We recently redesigned the downloads page to provide more information about each file available. The downloads are grouped into a section of files available directly from ZFIN and a section of files provided by external sources. Subgroups organized by data type make it easier to find the appropriate download. Files are listed in a table with a file description, including the file size and the number of records. Clicking on the file link displays its contents in the web browser. Alternatively, a download can be started immediately by clicking a button with the file format indicated. An additional link provides a header with details that describe each column in the file. Most files are tab-delimited with the exception of the GBrowse data files that use gff3 file format, a slightly extended tab-delimited standard format for exchanging genomic coordinate data.
To support access to the growing number of mutants in our database, we added two new files to the downloads collection, files for ‘All Genomic Features’ and ‘Genomic Features and their affected genes’. The ‘All Genomic Features’ file provides details about every allele and transgenic insertion recorded in ZFIN. The ‘Genomic Features and their affected genes’ file lists all the gene alleles for each feature, as well as ‘markers missing’ from deficiencies. All the download files hosted by ZFIN are updated nightly.
Reproducibility is one of the most basic tenets of scientific research. The common use of downloaded data in large-scale bioinformatics analyses has made it increasingly important to support access to snapshots of data versioned through time in addition to providing downloads of the most up-to-date data. ZFIN now supports downloads from archives that are captured on a daily basis. We strongly encourage users of any downloaded data sets to record and report the version and/or date stamp for the data and software they use. Without this information, reproducibility of results becomes virtually impossible due to changes in data and software over time.
CONCLUSION AND FUTURE DIRECTION
The zebrafish has emerged in recent years as a pre-eminent vertebrate model organism for studies aimed at understanding gene function and human disease. ZFIN is dedicated to supporting the research community in this ongoing effort. Among the most rapidly growing data types are mutants and transgenic lines. Recognizing this, ZFIN has begun collaborations with laboratories that produce large numbers of mutants and transgenics to ensure that core aspects of these important data are centrally located at ZFIN. We will continue to seek and support such collaborations in the future. To help researchers find mutants and transgenics, the Mutant/Morphant/Transgenic search interface was recently updated and backed with data mart infrastructure. Phenotype data associated with mutants and transgenics must be detailed and well structured to support mining human disease knowledge from the model organism annotation sets. We have increased the detail of our phenotype annotation methods, providing more expressive phenotype statements that will play an integral role in our ability to integrate phenotype data with data at other resources. In the future, we plan to expand our use of data marts to increase the speed and utility of all of our searches.
FUNDING
National Human Genome Research Institute (NHGRI) [HG002659, HG004838 and HG004834]; National Institutes of Health (NIH). Funding for open access charge: NHGRI HG002659; NIH (to M.W.).
Conflict of interest statement. None declared.




Comments