-
PDF
- Split View
-
Views
-
Cite
Cite
Sylvie Chevalier, Emeline Bouffartigues, Josselin Bodilis, Olivier Maillot, Olivier Lesouhaitier, Marc G. J. Feuilloley, Nicole Orange, Alain Dufour, Pierre Cornelis, Structure, function and regulation of Pseudomonas aeruginosa porins, FEMS Microbiology Reviews, Volume 41, Issue 5, September 2017, Pages 698–722, https://doi.org/10.1093/femsre/fux020
Close - Share Icon Share
Abstract
Pseudomonas aeruginosa is a Gram-negative bacterium belonging to the γ-proteobacteria. Like other members of the Pseudomonas genus, it is known for its metabolic versatility and its ability to colonize a wide range of ecological niches, such as rhizosphere, water environments and animal hosts, including humans where it can cause severe infections. Another particularity of P. aeruginosa is its high intrinsic resistance to antiseptics and antibiotics, which is partly due to its low outer membrane permeability. In contrast to Enterobacteria, pseudomonads do not possess general diffusion porins in their outer membrane, but rather express specific channel proteins for the uptake of different nutrients. The major outer membrane ‘porin’, OprF, has been extensively investigated, and displays structural, adhesion and signaling functions while its role in the diffusion of nutrients is still under discussion. Other porins include OprB and OprB2 for the diffusion of glucose, the two small outer membrane proteins OprG and OprH, and the two porins involved in phosphate/pyrophosphate uptake, OprP and OprO. The remaining nineteen porins belong to the so-called OprD (Occ) family, which is further split into two subfamilies termed OccD (8 members) and OccK (11 members). In the past years, a large amount of information concerning the structure, function and regulation of these porins has been published, justifying why an updated review is timely.
INTRODUCTION
Pseudomonas aeruginosa is the best known and investigated member of the genus Pseudomonas, the representatives of which are known for their high metabolic versatility. Pseudomonas syringae pathovars are important plant pathogens while P. aeruginosa causes infections in immunocompromised individuals and in cystic fibrosis (CF) patients (Goldberg 2000; Lyczak, Cannon and Pier 2000). Being a Gram-negative bacterium, P. aeruginosa has a cytoplasmic membrane with a symmetric phospholipid bilayer and an asymmetric outer membrane with a phospholipid inner face and a lipopolysaccharide outer layer, which generates a permeability barrier. The outer membrane of P. aeruginosa contains numerous proteins, including lipoproteins and channels (Remans et al. 2010). Exchange of nutrients across the outer membrane is orchestrated by β-barrel proteins producing water-filled diffusion channels, which give these membranes a molecular sieve-like appearance. These channels were first termed porins, given their pore shape and function, and were characterized by electron microscopy and conductance measurements in planar lipid membranes (Nakae 1976; Hancock, Decad and Nikaido 1979). Porins fold in the outer membrane as β-barrels made of antiparallel β-sheets with hydrophobic amino acids facing outward and hydrophilic residues inside the barrel and lining the constricted pore (Fernandez and Hancock 2012). This definition is still nowadays used in case of members of the Pseudomonas genus. However, in the case of Enterobacteriaceae, the term ‘porin’ was exclusively affected to large non-specific outer membrane β-barrel channels such as OmpF or OmpC (Nikaido 2003). Thus, two classes of outer membrane diffusion channels can be distinguished in Gram-negative bacteria: non-specific large general porins and substrate-specific channels (van den Berg 2012). Since Pseudomonas members do not display such large general porins, applying strictly this definition would result in the absence of porins. However, since numerous studies focusing on Pseudomonas members still use the term porin for outer membrane channels, it will be used throughout this review. We will therefore stick to the definition by Henderson et al. (2016) who proposes that ‘the outer membrane β-barrel proteins termed porins allow the passage of solutes or contribute to the envelope stability’. Pseudomonas aeruginosa is characterized by the very low permeability of its outer membrane, representing about 8% of that of Escherichia coli, which at least partly contributes to the high intrinsic and induced resistance to antibiotics (Hancock 1998). One of the reasons for this low permeability is the already mentioned absence of large general diffusion porins, such as OmpF and OmpC (Pratt et al. 1996). Another reason is that the OprD (Occ) family comprises 19 members (Hancock and Brinkman 2002; Tamber, Ochs and Hancock 2006; Liu et al. 2012a,b), which together are involved in the specific uptake of a wide range of small molecules of typically 200 Da or less (Eren et al. 2012). Crystal structures for 14 members of this family have been obtained, all of which show channels that are substantially narrower than those of the enterobacteria porins (Eren et al. 2012).
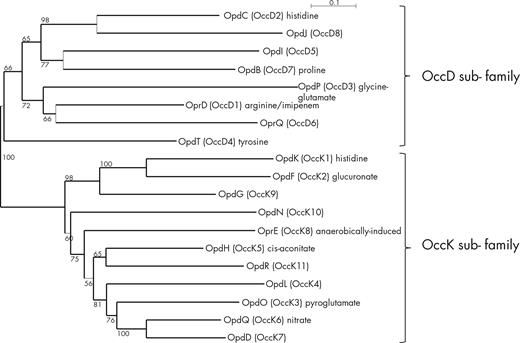
There are different families of porins, including the so-called structural porins of the OmpA family (Smith et al. 2007; Confer and Ayalew 2013), the small porins of the OmpW family (8 β-sheets) (Hong et al. 2006; Benz et al. 2015; Catel-Ferreira et al. 2016) and larger diffusion porins with 18 β-sheets (Hancock and Brinkman 2002). Still, larger channels with 22 anti-parallel β-sheets are present in the outer membrane, the TonB-dependent receptors for the uptake of siderophores, heme and organic sulfur molecules. They are energized by the TonB inner membrane protein which relays the proton motive force opening the gate to permit the passage of large molecules (Cornelis and Bodilis 2009). Other specialized channels are involved in the efflux of toxic molecules, including antibiotics, or participate in secretion systems, but are not going to be discussed in this review (Hancock and Brinkman 2002; Schweizer 2003). This review will therefore focus on the 26 porins of P. aeruginosa, including OprF, which is the major non-lipoprotein outer membrane protein, and the homolog of OmpA of E. coli. Given the many data that recently emerged on OprF, and in terms of its numerous important functions as well as the complex regulation of its expression, this review will put a particular emphasis on this amazing protein. The other porins described here are the two small OprG and OprH proteins, the two OprB glucose porins, the OprO and OprP phosphate porins and the members of the so-called OprD (Occ) family (Hancock and Brinkman 2002; Tamber, Ochs and Hancock 2006; Liu et al. 2012a,b). The 19 members of the OprD family which have been renamed Occ (outer membrane carboxylate channel) are phylogenetically split into two subfamilies, OccD being involved in the uptake of basic amino acids and the OccK for the uptake of negatively charged cyclic molecules (Tamber, Ochs and Hancock 2006; Eren et al.2012, 2013). However, despite the new nomenclature that takes into account the functions of these channels, most articles still use the old Opr- Opd- names. To help understanding, the double nomenclature will be used throughout this review.
GENOMIC CONTEXT AND EXPRESSION REGULATION
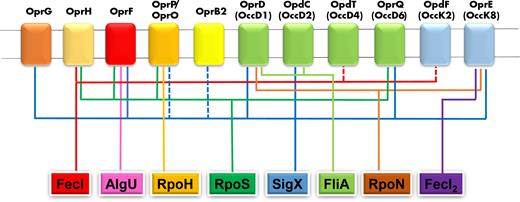
Most porin genes (16 out of 26) are transcribed as monocistronic mRNAs, which does not mean that they are not co-transcribed or co-regulated with other genes in the vicinity (Table 1). However, some outer membrane porin genes are clearly in an operonic structure as shown in Table 1 and in Fig. 1. Many regulators are involved in the control of porin genes expression, including (i) two-component systems where a sensor in the inner membrane detects a signal which is relayed by phosphorylation to a response regulator (Rodrigue et al. 2000; Zschiedrich, Keidel and Szurmant 2016); (ii) extracytoplasmic function (ECF) sigma factors, which are transcription factors that are involved in stress responses perceived outside the cytoplasm, and/or in the regulation of numerous genes encoding proteins having ECFs (Potvin, Sanschagrin and Levesque 2008; Llamas et al. 2014; Schulz et al. 2015). The involvement of the different types of regulators in the expression of porin genes will be developed further in this review.
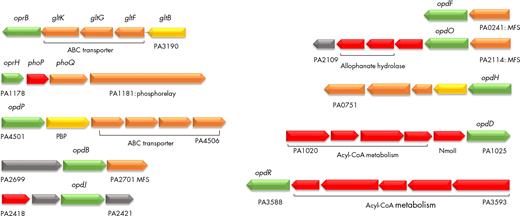
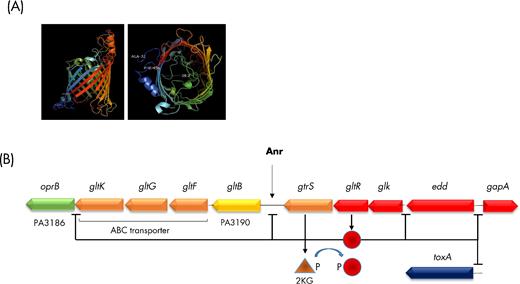
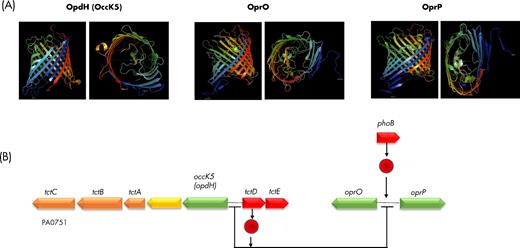
Porin genes contained in operons. Operons including porin-encoding genes from P. aeruginosa PAO1 (pseudomonas.com). Predicted localization of the encoded proteins is indicated by colors as in Pseudomonas.com (green: outer membrane; orange: cytoplasmic membrane; red: cytoplasmic; yellow: periplasmic; gray: unknown). ABC: ATP-binding cassette; PBP: probable binding protein component of ABC transporter; MFS: major facilitator superfamily transporter; NmoII: type II nitronate monooxygenase (Salvi et al. 2014).
List of porin genes and alternative porin name designation, PAO1 gene number, PA14 gene number, molecular mass, direction (Dir.) of transcription (>forward strand, <reverse strand), operonic structure, possible function, antibiotic (ATB) transport, regulators controlling the porin expression, number of orthologs, orthologs group and associated transporter genes (http://pseudomonas.com; Winsor et al. 2016).
| Gene . | PAO1 n° . | PA14 n° . | Mass (kDa) . | Dir. . | Operon . | Function . | ATB transport . | Regulators . | Sigma factorsa . | Orthologs . | Ortholog group (*) . | Transporter-associated . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| oprF | PA1777 | PA14_41570 | 37.6 | > | Nob | Structuralc | AmpR | AlgU1,3, SigX*1, RpoS1 | 48 | POG003618 (572) | ||
| oprB | PA3186 | PA14_23030 | 50.8 | < | PA3190-3186 | Glucose uptake | Anr, GltR | RpoS2,3 | 38 | POG002430 (735) | PA3189-3187 (ABC) | |
| oprB2 | PA2291 | PA14_34960 | 50.5 | > | No | Glucose uptake | ? | (SigX) | 37 | POG002430 (735) | ||
| oprG | PA4067 | PA14_11270 | 25.2 | > | No | Fe2+ uptake? | Anr | SigX1,2,3, FecI2 | 48 | POG001618 (570) | ||
| oprH | PA1178 | PA14_49200 | 21.6 | > | oprH-phoP-PhoQ | OM stabilization | PhoP-PhoQ, BrlR, BqsR/CarR | RpoS1,2, FecI1 | 41 | POG001136 (469) | ||
| oprP | PA3279 | PA14_21620 | 48.2 | < | No | Phosphate | PhoB, TctD | RpoH1,2, RpoS1, (SigX) | 28 | POG002334 (719) | ||
| oprO | PA3280 | PA14_21610 | 47.8 | < | No | Pyrophosphate | PhoB, TctD | RpoH1, RpoS1, (SigX) | 25 | POG002334 (719) | ||
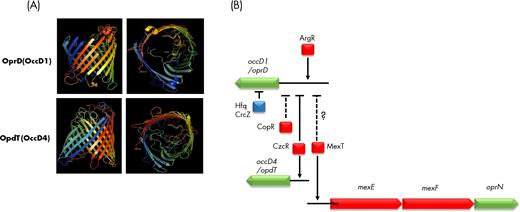
| oprD (occD1) | PA0958 | PA14_51880 | 48.4 | < | No | Arginine | Imipenem/ meropenem | ArgR, CzcR | RpoN1,3, SigX1,3, FliA1 | 37 | POG000187 (1656) | |
| opdC (occD2) | PA0162 | PA14_02020 | 48.9 | > | No | Histidine, arginine | SigX1,2,3, FliA1 | 29 | POG000160 (459) | |||
| opdP (occD3) | PA4503 | PA14_58410 | 53 | > | PA4501-4506 | Gly-Glu, arginine | RpoN2,3 | 38 | POG004088 (516) | ABC transporter | ||
| opdT (occD4) | PA2505 | PA14_32270 | 49.8 | < | No | Tyrosine | ? | (FecI) | 49 | POG000187 (1656) | ||
| opdI (occD5) | PA0189 | PA14_02370 | 48.9 | < | No | Arginine | ? | ? | 14 | POG000187 (1656) | ||
| oprQ (occD6) | PA2760 | PA14_28400 | 46.9 | > | No | Arginine | MexT | RpoS1, SigX1 | 48 | POG002810 (552) | ||
| opdB (occD7) | PA2700 | PA14_29220 | 47.3 | > | PA2699-2701 | Proline, arginine | ? | 23 | POG000187 (1656) | MFS | ||
| opdJ (occD8) | PA2420 | PA14_33410 | 51.3 | > | PA2418-2421 | Arginine | ? | ? | 14 | POG003114 (312) | PA2419 is an hydrolase | |
| opdK (occK1) | PA4898 | PA14_64720 | 45.8 | < | No | Vanillate, benzoate | Carbenicillin, cefoxitin, tetracyclin, temocillin | ? | ? | 26 | POG000239 (1215) | |
| opdF (occK2) | PA0240 | PA14_02980 | 46.1 | < | PA0241 | Glucuronate | Carbenicillin, cefoxitin, gentamycin, temocillin | ? | (FecI) | 68 | POG000239 (1215) | MFS |
| opdO (occK3) | PA2113 | PA14_37260 | 44.3 | < | PA2114-2110 | Pyroglutamate | Cefotaxime | ? | ? | 14 | POG003356 (336) | PA2114 is an MFS |
| opdL (occK4) | PA4137 | PA14_10440 | 46.04 | > | No | Phenylacetate | ? | 26 | POG001562 (382) | |||
| opdH (occK5) | PA0755 | PA14_54520 | 47 | < | PA0755-0751tctCBA | Cis-aconitate, tri-carboxylates | TctD | ? | 39 | POG000729 (882) | (tctABC)Tri-carboxylate transport | |
| opdQ (occK6) | PA3038 | PA14_24790 | 46.7 | > | No | NarXL | ? | 28 | POG002549 (380) | |||
| opdD (occK7) | PA1025 | PA14_51070 | 46.5 | > | PA1020-1025 | Meropenem | ? | ? | 24 | POG000987 (301) | Acyl-CoA dehydrogenase, PA1024 nitropropane dioxygenase | |
| oprE (occK8) | PA0291 | PA14_03800 | 49.7 | > | No | IHF, OxyR | SigX1, RpoN1,2, FecI21 | 53 | POG000288 (540) | |||
| opdG (occK9) | PA2213 | PA14_36090 | 45.4 | > | No | PA2206 | ? | 23 | POG000239 (1215) | |||
| opdN (occK10) | PA4179 | PA14_09850 | 47.8 | > | No | ? | ? | 26 | POG000729 (882) | |||
| opdR (occK11) | PA3588 | PA14_17890 | 45.7 | < | PA3593-3588 | ? | 13 | POG002082 (306) | Acyl-coA metabolism |
| Gene . | PAO1 n° . | PA14 n° . | Mass (kDa) . | Dir. . | Operon . | Function . | ATB transport . | Regulators . | Sigma factorsa . | Orthologs . | Ortholog group (*) . | Transporter-associated . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| oprF | PA1777 | PA14_41570 | 37.6 | > | Nob | Structuralc | AmpR | AlgU1,3, SigX*1, RpoS1 | 48 | POG003618 (572) | ||
| oprB | PA3186 | PA14_23030 | 50.8 | < | PA3190-3186 | Glucose uptake | Anr, GltR | RpoS2,3 | 38 | POG002430 (735) | PA3189-3187 (ABC) | |
| oprB2 | PA2291 | PA14_34960 | 50.5 | > | No | Glucose uptake | ? | (SigX) | 37 | POG002430 (735) | ||
| oprG | PA4067 | PA14_11270 | 25.2 | > | No | Fe2+ uptake? | Anr | SigX1,2,3, FecI2 | 48 | POG001618 (570) | ||
| oprH | PA1178 | PA14_49200 | 21.6 | > | oprH-phoP-PhoQ | OM stabilization | PhoP-PhoQ, BrlR, BqsR/CarR | RpoS1,2, FecI1 | 41 | POG001136 (469) | ||
| oprP | PA3279 | PA14_21620 | 48.2 | < | No | Phosphate | PhoB, TctD | RpoH1,2, RpoS1, (SigX) | 28 | POG002334 (719) | ||
| oprO | PA3280 | PA14_21610 | 47.8 | < | No | Pyrophosphate | PhoB, TctD | RpoH1, RpoS1, (SigX) | 25 | POG002334 (719) | ||
| oprD (occD1) | PA0958 | PA14_51880 | 48.4 | < | No | Arginine | Imipenem/ meropenem | ArgR, CzcR | RpoN1,3, SigX1,3, FliA1 | 37 | POG000187 (1656) | |
| opdC (occD2) | PA0162 | PA14_02020 | 48.9 | > | No | Histidine, arginine | SigX1,2,3, FliA1 | 29 | POG000160 (459) | |||
| opdP (occD3) | PA4503 | PA14_58410 | 53 | > | PA4501-4506 | Gly-Glu, arginine | RpoN2,3 | 38 | POG004088 (516) | ABC transporter | ||
| opdT (occD4) | PA2505 | PA14_32270 | 49.8 | < | No | Tyrosine | ? | (FecI) | 49 | POG000187 (1656) | ||
| opdI (occD5) | PA0189 | PA14_02370 | 48.9 | < | No | Arginine | ? | ? | 14 | POG000187 (1656) | ||
| oprQ (occD6) | PA2760 | PA14_28400 | 46.9 | > | No | Arginine | MexT | RpoS1, SigX1 | 48 | POG002810 (552) | ||
| opdB (occD7) | PA2700 | PA14_29220 | 47.3 | > | PA2699-2701 | Proline, arginine | ? | 23 | POG000187 (1656) | MFS | ||
| opdJ (occD8) | PA2420 | PA14_33410 | 51.3 | > | PA2418-2421 | Arginine | ? | ? | 14 | POG003114 (312) | PA2419 is an hydrolase | |
| opdK (occK1) | PA4898 | PA14_64720 | 45.8 | < | No | Vanillate, benzoate | Carbenicillin, cefoxitin, tetracyclin, temocillin | ? | ? | 26 | POG000239 (1215) | |
| opdF (occK2) | PA0240 | PA14_02980 | 46.1 | < | PA0241 | Glucuronate | Carbenicillin, cefoxitin, gentamycin, temocillin | ? | (FecI) | 68 | POG000239 (1215) | MFS |
| opdO (occK3) | PA2113 | PA14_37260 | 44.3 | < | PA2114-2110 | Pyroglutamate | Cefotaxime | ? | ? | 14 | POG003356 (336) | PA2114 is an MFS |
| opdL (occK4) | PA4137 | PA14_10440 | 46.04 | > | No | Phenylacetate | ? | 26 | POG001562 (382) | |||
| opdH (occK5) | PA0755 | PA14_54520 | 47 | < | PA0755-0751tctCBA | Cis-aconitate, tri-carboxylates | TctD | ? | 39 | POG000729 (882) | (tctABC)Tri-carboxylate transport | |
| opdQ (occK6) | PA3038 | PA14_24790 | 46.7 | > | No | NarXL | ? | 28 | POG002549 (380) | |||
| opdD (occK7) | PA1025 | PA14_51070 | 46.5 | > | PA1020-1025 | Meropenem | ? | ? | 24 | POG000987 (301) | Acyl-CoA dehydrogenase, PA1024 nitropropane dioxygenase | |
| oprE (occK8) | PA0291 | PA14_03800 | 49.7 | > | No | IHF, OxyR | SigX1, RpoN1,2, FecI21 | 53 | POG000288 (540) | |||
| opdG (occK9) | PA2213 | PA14_36090 | 45.4 | > | No | PA2206 | ? | 23 | POG000239 (1215) | |||
| opdN (occK10) | PA4179 | PA14_09850 | 47.8 | > | No | ? | ? | 26 | POG000729 (882) | |||
| opdR (occK11) | PA3588 | PA14_17890 | 45.7 | < | PA3593-3588 | ? | 13 | POG002082 (306) | Acyl-coA metabolism |
According to ChIP data (Schulz et al.2015). Under brackets are shown sigma factors that have not been considered statistically significant but interactions were positive in the two repetitions.
The genomic context of oprF is detailed in the text and in Fig. 2.
The multiple functions of OprF are detailed in the text.
Involvement of SigX in oprF transcription is discussed in the text.
(*) Number of members.
1ChIP, 2Motif, 3RNA seq (according to Schulz et al.2015).
List of porin genes and alternative porin name designation, PAO1 gene number, PA14 gene number, molecular mass, direction (Dir.) of transcription (>forward strand, <reverse strand), operonic structure, possible function, antibiotic (ATB) transport, regulators controlling the porin expression, number of orthologs, orthologs group and associated transporter genes (http://pseudomonas.com; Winsor et al. 2016).
| Gene . | PAO1 n° . | PA14 n° . | Mass (kDa) . | Dir. . | Operon . | Function . | ATB transport . | Regulators . | Sigma factorsa . | Orthologs . | Ortholog group (*) . | Transporter-associated . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| oprF | PA1777 | PA14_41570 | 37.6 | > | Nob | Structuralc | AmpR | AlgU1,3, SigX*1, RpoS1 | 48 | POG003618 (572) | ||
| oprB | PA3186 | PA14_23030 | 50.8 | < | PA3190-3186 | Glucose uptake | Anr, GltR | RpoS2,3 | 38 | POG002430 (735) | PA3189-3187 (ABC) | |
| oprB2 | PA2291 | PA14_34960 | 50.5 | > | No | Glucose uptake | ? | (SigX) | 37 | POG002430 (735) | ||
| oprG | PA4067 | PA14_11270 | 25.2 | > | No | Fe2+ uptake? | Anr | SigX1,2,3, FecI2 | 48 | POG001618 (570) | ||
| oprH | PA1178 | PA14_49200 | 21.6 | > | oprH-phoP-PhoQ | OM stabilization | PhoP-PhoQ, BrlR, BqsR/CarR | RpoS1,2, FecI1 | 41 | POG001136 (469) | ||
| oprP | PA3279 | PA14_21620 | 48.2 | < | No | Phosphate | PhoB, TctD | RpoH1,2, RpoS1, (SigX) | 28 | POG002334 (719) | ||
| oprO | PA3280 | PA14_21610 | 47.8 | < | No | Pyrophosphate | PhoB, TctD | RpoH1, RpoS1, (SigX) | 25 | POG002334 (719) | ||
| oprD (occD1) | PA0958 | PA14_51880 | 48.4 | < | No | Arginine | Imipenem/ meropenem | ArgR, CzcR | RpoN1,3, SigX1,3, FliA1 | 37 | POG000187 (1656) | |
| opdC (occD2) | PA0162 | PA14_02020 | 48.9 | > | No | Histidine, arginine | SigX1,2,3, FliA1 | 29 | POG000160 (459) | |||
| opdP (occD3) | PA4503 | PA14_58410 | 53 | > | PA4501-4506 | Gly-Glu, arginine | RpoN2,3 | 38 | POG004088 (516) | ABC transporter | ||
| opdT (occD4) | PA2505 | PA14_32270 | 49.8 | < | No | Tyrosine | ? | (FecI) | 49 | POG000187 (1656) | ||
| opdI (occD5) | PA0189 | PA14_02370 | 48.9 | < | No | Arginine | ? | ? | 14 | POG000187 (1656) | ||
| oprQ (occD6) | PA2760 | PA14_28400 | 46.9 | > | No | Arginine | MexT | RpoS1, SigX1 | 48 | POG002810 (552) | ||
| opdB (occD7) | PA2700 | PA14_29220 | 47.3 | > | PA2699-2701 | Proline, arginine | ? | 23 | POG000187 (1656) | MFS | ||
| opdJ (occD8) | PA2420 | PA14_33410 | 51.3 | > | PA2418-2421 | Arginine | ? | ? | 14 | POG003114 (312) | PA2419 is an hydrolase | |
| opdK (occK1) | PA4898 | PA14_64720 | 45.8 | < | No | Vanillate, benzoate | Carbenicillin, cefoxitin, tetracyclin, temocillin | ? | ? | 26 | POG000239 (1215) | |
| opdF (occK2) | PA0240 | PA14_02980 | 46.1 | < | PA0241 | Glucuronate | Carbenicillin, cefoxitin, gentamycin, temocillin | ? | (FecI) | 68 | POG000239 (1215) | MFS |
| opdO (occK3) | PA2113 | PA14_37260 | 44.3 | < | PA2114-2110 | Pyroglutamate | Cefotaxime | ? | ? | 14 | POG003356 (336) | PA2114 is an MFS |
| opdL (occK4) | PA4137 | PA14_10440 | 46.04 | > | No | Phenylacetate | ? | 26 | POG001562 (382) | |||
| opdH (occK5) | PA0755 | PA14_54520 | 47 | < | PA0755-0751tctCBA | Cis-aconitate, tri-carboxylates | TctD | ? | 39 | POG000729 (882) | (tctABC)Tri-carboxylate transport | |
| opdQ (occK6) | PA3038 | PA14_24790 | 46.7 | > | No | NarXL | ? | 28 | POG002549 (380) | |||
| opdD (occK7) | PA1025 | PA14_51070 | 46.5 | > | PA1020-1025 | Meropenem | ? | ? | 24 | POG000987 (301) | Acyl-CoA dehydrogenase, PA1024 nitropropane dioxygenase | |
| oprE (occK8) | PA0291 | PA14_03800 | 49.7 | > | No | IHF, OxyR | SigX1, RpoN1,2, FecI21 | 53 | POG000288 (540) | |||
| opdG (occK9) | PA2213 | PA14_36090 | 45.4 | > | No | PA2206 | ? | 23 | POG000239 (1215) | |||
| opdN (occK10) | PA4179 | PA14_09850 | 47.8 | > | No | ? | ? | 26 | POG000729 (882) | |||
| opdR (occK11) | PA3588 | PA14_17890 | 45.7 | < | PA3593-3588 | ? | 13 | POG002082 (306) | Acyl-coA metabolism |
| Gene . | PAO1 n° . | PA14 n° . | Mass (kDa) . | Dir. . | Operon . | Function . | ATB transport . | Regulators . | Sigma factorsa . | Orthologs . | Ortholog group (*) . | Transporter-associated . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| oprF | PA1777 | PA14_41570 | 37.6 | > | Nob | Structuralc | AmpR | AlgU1,3, SigX*1, RpoS1 | 48 | POG003618 (572) | ||
| oprB | PA3186 | PA14_23030 | 50.8 | < | PA3190-3186 | Glucose uptake | Anr, GltR | RpoS2,3 | 38 | POG002430 (735) | PA3189-3187 (ABC) | |
| oprB2 | PA2291 | PA14_34960 | 50.5 | > | No | Glucose uptake | ? | (SigX) | 37 | POG002430 (735) | ||
| oprG | PA4067 | PA14_11270 | 25.2 | > | No | Fe2+ uptake? | Anr | SigX1,2,3, FecI2 | 48 | POG001618 (570) | ||
| oprH | PA1178 | PA14_49200 | 21.6 | > | oprH-phoP-PhoQ | OM stabilization | PhoP-PhoQ, BrlR, BqsR/CarR | RpoS1,2, FecI1 | 41 | POG001136 (469) | ||
| oprP | PA3279 | PA14_21620 | 48.2 | < | No | Phosphate | PhoB, TctD | RpoH1,2, RpoS1, (SigX) | 28 | POG002334 (719) | ||
| oprO | PA3280 | PA14_21610 | 47.8 | < | No | Pyrophosphate | PhoB, TctD | RpoH1, RpoS1, (SigX) | 25 | POG002334 (719) | ||
| oprD (occD1) | PA0958 | PA14_51880 | 48.4 | < | No | Arginine | Imipenem/ meropenem | ArgR, CzcR | RpoN1,3, SigX1,3, FliA1 | 37 | POG000187 (1656) | |
| opdC (occD2) | PA0162 | PA14_02020 | 48.9 | > | No | Histidine, arginine | SigX1,2,3, FliA1 | 29 | POG000160 (459) | |||
| opdP (occD3) | PA4503 | PA14_58410 | 53 | > | PA4501-4506 | Gly-Glu, arginine | RpoN2,3 | 38 | POG004088 (516) | ABC transporter | ||
| opdT (occD4) | PA2505 | PA14_32270 | 49.8 | < | No | Tyrosine | ? | (FecI) | 49 | POG000187 (1656) | ||
| opdI (occD5) | PA0189 | PA14_02370 | 48.9 | < | No | Arginine | ? | ? | 14 | POG000187 (1656) | ||
| oprQ (occD6) | PA2760 | PA14_28400 | 46.9 | > | No | Arginine | MexT | RpoS1, SigX1 | 48 | POG002810 (552) | ||
| opdB (occD7) | PA2700 | PA14_29220 | 47.3 | > | PA2699-2701 | Proline, arginine | ? | 23 | POG000187 (1656) | MFS | ||
| opdJ (occD8) | PA2420 | PA14_33410 | 51.3 | > | PA2418-2421 | Arginine | ? | ? | 14 | POG003114 (312) | PA2419 is an hydrolase | |
| opdK (occK1) | PA4898 | PA14_64720 | 45.8 | < | No | Vanillate, benzoate | Carbenicillin, cefoxitin, tetracyclin, temocillin | ? | ? | 26 | POG000239 (1215) | |
| opdF (occK2) | PA0240 | PA14_02980 | 46.1 | < | PA0241 | Glucuronate | Carbenicillin, cefoxitin, gentamycin, temocillin | ? | (FecI) | 68 | POG000239 (1215) | MFS |
| opdO (occK3) | PA2113 | PA14_37260 | 44.3 | < | PA2114-2110 | Pyroglutamate | Cefotaxime | ? | ? | 14 | POG003356 (336) | PA2114 is an MFS |
| opdL (occK4) | PA4137 | PA14_10440 | 46.04 | > | No | Phenylacetate | ? | 26 | POG001562 (382) | |||
| opdH (occK5) | PA0755 | PA14_54520 | 47 | < | PA0755-0751tctCBA | Cis-aconitate, tri-carboxylates | TctD | ? | 39 | POG000729 (882) | (tctABC)Tri-carboxylate transport | |
| opdQ (occK6) | PA3038 | PA14_24790 | 46.7 | > | No | NarXL | ? | 28 | POG002549 (380) | |||
| opdD (occK7) | PA1025 | PA14_51070 | 46.5 | > | PA1020-1025 | Meropenem | ? | ? | 24 | POG000987 (301) | Acyl-CoA dehydrogenase, PA1024 nitropropane dioxygenase | |
| oprE (occK8) | PA0291 | PA14_03800 | 49.7 | > | No | IHF, OxyR | SigX1, RpoN1,2, FecI21 | 53 | POG000288 (540) | |||
| opdG (occK9) | PA2213 | PA14_36090 | 45.4 | > | No | PA2206 | ? | 23 | POG000239 (1215) | |||
| opdN (occK10) | PA4179 | PA14_09850 | 47.8 | > | No | ? | ? | 26 | POG000729 (882) | |||
| opdR (occK11) | PA3588 | PA14_17890 | 45.7 | < | PA3593-3588 | ? | 13 | POG002082 (306) | Acyl-coA metabolism |
According to ChIP data (Schulz et al.2015). Under brackets are shown sigma factors that have not been considered statistically significant but interactions were positive in the two repetitions.
The genomic context of oprF is detailed in the text and in Fig. 2.
The multiple functions of OprF are detailed in the text.
Involvement of SigX in oprF transcription is discussed in the text.
(*) Number of members.
1ChIP, 2Motif, 3RNA seq (according to Schulz et al.2015).
STRUCTURAL CHARACTERISTICS OF PSEUDOMONAS AERUGINOSA OUTER MEMBRANE PORINS
Most porins from P. aeruginosa have a molecular mass ranging from 45.4 kDa (OpdG/OccK9) to 53 kDa (OpdP/OccD3), except the two smaller OprG (25.2kDa) and OprH (21.6 kDa) and the OprF porin (37.6 kDa) (Table 1). The structure of 16 porins has been determined (Fig. S1,Supporting Information; Table 1), including the small β-barrel porin OprG with only 8 β-sheets (Kucharska et al.2015, 2016), while the porins belonging to the OprD/Occ family all have 18 β-sheets. The structure of the OprF porin is still a matter of debate, since full-length OprF resists crystallographic efforts and the link between its functions and tertiary or quaternary structures remains controversial and will be discussed further. The OprD/Occ family comprise 19 members divided into two subfamilies: the OccD (8 members) and the OccK (11 members) (Tamber, Ochs and Hancock 2006; Eren et al. 2012). The structures of OccD1-3 and OccK1-OccK6 have been determined (Fig. S1) (Eren et al.2012, 2013). The porins of the OccD family have a smaller pore compared to those of the OccK subfamily, but all have a basic ladder (row of Arg and Lys residues) in the barrel wall, the side chains pointing to the lumen (Eren et al. 2012, 2013). The OccD porins have a more dynamic channel which can be closed or open while the OccK porins have a more rigid channel (Eren et al.2012, 2013).
OprF, THE MAJOR AND MULTIFARIOUS PORIN OF PSEUDOMONAS AERUGINOSA
As the most abundant non-lipoprotein outer membrane protein in P. aeruginosa, OprF has been the object of numerous research works since 1979 (Hancock and Carey 1979; Hancock, Decad and Nikaido 1979). The most recent reviews dealing with OprF focused on two specific aspects, folding pathways (Sugawara, Nagano and Nikaido 2012) and involvement in pathogenesis (Krishnan and Prasadarao 2012), but a comprehensive review on this protein is missing. We therefore devoted a large part of the present review on OprF, which is justified by the large amount of available data, the variety of functions in which OprF is involved, and recent new findings, for example in the regulation of the oprF gene.
The elusive structure of OprF
OprF of P. aeruginosa is homologous to the outer membrane protein A (OmpA) of Escherichia coli, and these two proteins are the best studied members of the OmpA protein family. The structures of OmpA and OprF were reviewed and compared in 2012 (Reusch 2012; Sugawara, Nagano and Nikaido 2012). Briefly, OprF (326 residues) folds into three domains: the crystallized N-terminal eight-stranded β-barrel located in the outer membrane (Brinkman, Bains and Hancock 2000; Reusch 2012) (Fig. 2A, Fig. S1), a cysteine-rich linker that may be partly surface exposed (Hancock and Carey 1979; Hancock, Decad and Nikaido 1979; Bodilis et al. 2004) and the C-terminal part containing α-helixes and/or β-strands (Sugawara, Nagano and Nikaido 2012). As OmpA, OprF generates two distinct conformers corresponding to a closed channel (the most abundant form) and a rare (<5% of the conformers) open channel (Sugawara and Nikaido 1994). The closed conformer contains the two domains mentioned above (the N-terminal eight-stranded β-barrel and the C-terminal periplasmic globular domain), whereas the open form folds as a single domain protein with a larger number of transmembrane β-strands (14 to 16) (Sugawara et al. 2006; Sugawara, Nagano and Nikaido 2012). The OprF channels mainly exist in weakly conductive subconformations and switch to the fully open state for a short time only (Nestorovich et al. 2006), thereby contributing to the low permeability of P. aeruginosa outer membrane reported earlier (Bellido et al. 1992). Interestingly, the channel conductance can be modulated in function of the bacterial growth temperature, with 80 pS and 250 pS measured when bacteria were grown at 17°C or 37°C, respectively (Jaouen et al. 2004). While the small channel size is consistent with the predicted structure of the crystalized N-terminal domain, the largest one remains controversial, and has been attributed either to a folding of OprF as a large single domain or to the oligomerization of three subunits folding into three small channels of about 80 pS each (Jaouen et al. 2004; Sugawara et al. 2006). Remarkably, the recent discovery of unfolded OmpA monomers and oligomers that mimic amyloid fibers structures sheds new light into OmpA, and possibly also into OprF structures and associated functions (Wang et al. 2013; Danoff and Fleming 2015). It is interesting to mention that the largest OprF conformer proportion in the outer membrane increases in a mutant that does not make the periplasmic chaperone Skp, suggesting that in the absence of this chaperone, OprF could be misfolded like in the case of OmpA (Sugawara, Nagano and Nikaido 2010, 2012).
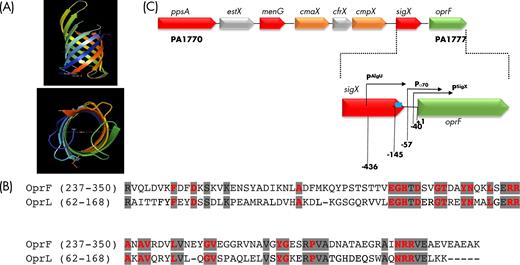
oprF genomic context and C-terminal protein similarity with OprL. (A) Schematic representation (PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC) viewed from the side (top) and from the extracellular environment (bottom) of the N-terminal part of OprF based on the X-ray crystal structure (PDB 4RLC). (B) Sequence (amino-acids) conservation between OprF and OprL. Conserved residues between the two proteins are shaded in gray. Peptidoglycan-binding residues are indicated in red (Cascales and Lloubes 2004). (C) Genomic locus including oprF and the seven genes upstream genes and localization of oprF promoter regions. The transcriptional initiation sites of the three promoters lying upstream of the oprF gene (PSigX, Pσ70, PAlgU), corresponding to SigX-, σ70- and AlgU-dependent promoters, respectively, are indicated by arrows. Their positions are indicated relative to the translational initiation start of oprF (+1). Position of AmpR putative binding site is indicated (blue square).
OprF as a channel
OprF was first considered to function as a non-specific aqueous channel, allowing the passage of ions and low molecular mass sugars (Bellido et al. 1992), but it has also been suggested to allow the passage of toluene since a mutant not expressing oprF is more tolerant to toluene (Li et al. 1995). OprF has also been suggested to allow the diffusion of iron-chelated non-cognate siderophores (Meyer 1992). An interesting observation is the increased levels of OprF in P. aeruginosa cells grown under anaerobic conditions in the presence of nitrate, suggesting a possible involvement of OprF in the diffusion of nitrates and nitrites (Yoon et al. 2002).
OprF involvement in maintaining the outer membrane integrity
OprF plays a structural role, contributing to maintenance of the cell shape, especially under low osmolarity conditions, since the C-terminal part contains a peptidoglycan binding domain, which anchors the outer membrane to the peptidoglycan layer (Gotoh et al. 1989; Rawling, Brinkman and Hancock 1998). OprF mutants lacking various portions of the C-terminal part confirmed that the N-terminal 164 amino acids are sufficient for protein production and membrane insertion, while the C-terminal part is needed for stable interaction with peptidoglycan (Rawling, Brinkman and Hancock 1998). The cells expressing C-terminally truncated OprF were both sensitive to low osmolarity and their cell length was reduced (Rawling, Brinkman and Hancock 1998). A comparison with OprL, the peptidoglycan-associated lipoprotein (PAL) (Lim et al. 1997), reveals the presence of remarkably conserved residues in the C-terminal part of both proteins (Fig. 2B). These residues are also conserved in the different PAL homologs in Gram-negative bacteria and are involved in the association with peptidoglycan or with TolA, an inner membrane protein which, together with TolC, TolQ and TolR, forms the Tol-PAL complex that insures the outer membrane integrity (Journet et al. 1999; Cascales et al. 2002; Cascales and Lloubes 2004). In E. coli, the PAL lipoprotein can also dimerize and interact with OmpA (Cascales et al. 2002). Since OprF belongs to the OmpA family, and because the critical residues for the interactions with peptidoglycan and Tol proteins are present in its C-terminal part, it is reasonable to suggest that OprF could either dimerize or interact with OprL, TolA, TolB and peptidoglycan, in line with the published evidence of its role in maintaining cell integrity under some environmental stresses (Rawling, Brinkman and Hancock 1998). In a study aiming at probing the protein interaction network of P. aeruginosa PAO1 by in vivo covalently linking interacting protein partners, OprF was confirmed to form homodimers and to interact with OprL (Navare et al. 2015). OprF and OprL were found to interact via their C-terminal part containing the conserved residues mentioned above. The small and abundant OprI lipoprotein (Cornelis et al. 1989) was also shown to interact with both OprF and OprL, suggesting the existence of ternary OprF-OprL-OprI complexes involved in cell shape and outer membrane stability. Four other OprF partners were identified: the translation initiation factor IF-2 encoded by the infB gene and the PA1041, PA1522 and PA1964 proteins of unknown functions. Of these four proteins, only PA1041 is predicted to be an OmpA-like outer membrane lipoprotein (Remans et al. 2010; Navare et al. 2015; Winsor et al. 2016), but the relevance of these interactions remains unknown. Finally, it is worth mentioning that these conserved PAL motifs are not present in the other P. aeruginosa porins (results not shown).
OprF and biofilm formation
Pseudomonas aeruginosa forms thick biofilms (defined as microbial communities of sessile cells embedded into a matrix of extracellular polymeric substances that they have produced) under anaerobic conditions in the presence of nitrate, which is accompanied by higher levels of OprF in the outer membrane (Hassett et al. 2002; Yoon et al. 2002). Furthermore, an oprF-negative mutant forms very poor biofilms under these anaerobic conditions and the cells lack nitrite reductase activity, making an interesting link with the previous suggestion that OprF could be involved in the diffusion of nitrates/nitrites (Hassett et al. 2002; Yoon et al. 2002). In line with these results, it was demonstrated that hypoxic conditions, such as those encountered in the CF lung, favor a higher expression of oprF (Hogardt and Heesemann 2010; Eichner et al. 2014). In artificial sputum medium (ASM), mimicking the conditions in the CF lung, Sriramulu et al. (2005) found that OprF is needed for the formation of microcolonies and that the high levels of amino-acids present in this medium favor high OprF production levels, whereas low OprF levels were observed in ASM without added amino acids. However, in LB medium under microaerobic conditions, but in the absence of additional nitrate, Bouffartigues et al. (2015) showed that an oprF-negative mutant forms aggregates in liquid medium accompanied by higher levels of extracellular Pel polysaccharides, and more strongly attached biofilm, in stark contrast to the anaerobic growth conditions mentioned above. These phenotypes could partly be the result of increased levels of cyclic-di-GMP intracellular levels, which are known to favor biofilm formation (Bouffartigues et al. 2015). These contrasting results could be explained by the different conditions used (anaerobic vs aerobic), different media (LB vs ASM) and the presence or absence of nitrate.
OprF function in binding and adhesion to mammalian cells
Adhesion of P. aeruginosa to human lung alveolar epithelial cells has been found to be, at least partly, mediated by OprF since an oprF-negative mutant had its binding capacity reduced by about 60% while pre-incubation of epithelial cells with purified OprF or with a monoclonal antibody against OprF also reduced attachment (Azghani et al. 2002). Likewise, OprF has been found to be involved in the binding to human middle ear epithelial cells, probably facilitating the invasion of P. aeruginosa in otitis media (Mittal et al. 2014). In chronic suppurative otitis media caused by P. aeruginosa, actin rearrangement occurs due to phosphorylation of protein kinase C (PKC) and OprF was found to be necessary for PKC activation (Mittal et al. 2016). In a separate study, an oprF mutant of PAO1 showed a drastic reduction of adherence to rat glial cells and to Caco2/TC7 cells while wild-type adhesion level was restored upon complementation with the oprF gene in trans (Fito-Boncompte et al. 2011). OprF has also been found to interact with the lectin LecB, which is exposed at the surface of P. aeruginosa cells and both proteins contribute to the hemaglutination of erythrocytes (Funken et al. 2012).
OprF involvement in outer membrane vesicle biogenesis and functions
Outer membrane vesicles (OMVs) are nanostructures (20–300 nm in diameter) produced by almost all Gram-negative bacteria. They are spherical vesicles delimited by a bilayer membrane originating from the bacterial outer membrane. OMV membrane is therefore made of an outer lipopolysaccharide (LPS) leaflet, an inner phospholipid leaflet and outer membrane proteins. OMVs were also shown to contain bacterial periplasmic components (proteins and cell wall components), proteins of the bacterial inner membrane, cytoplasmic proteins, DNA and RNA, ions, metabolites and signaling molecules (Kulp and Kuehn 2010; Kim et al. 2015; Pathirana and Kaparakis-Liaskos 2016). They are produced by planktonic bacteria, but also by bacteria in biofilms and can thus be considered as components of biofilm matrixes (Schooling and Beveridge 2006).
Several general models of events leading to outer membrane budding and OMV biogenesis were proposed, including the loss of links between the outer membrane and the peptidoglycan layer in regions where vesiculation will take place (Kulp and Kuehn 2010). Since OprF and the lipoproteins OprL and OprI tether the outer membrane to peptidoglycan, their roles in P. aeruginosa PA14 OMV biogenesis were investigated and the absence of OprF led to OMV amounts increased by about 8-fold (Wessel et al. 2013). The PQS quorum-sensing signaling molecule is hydrophobic and associates with LPS in the external leaflet of the outer membrane, causing the outer leaflet to expand relative to the inner leaflet, yielding OMV budding from the bacterial surface (Schertzer and Whiteley 2012). In the oprF mutant of PA14, the higher OMV biogenesis level was shown to result from an increase in PQS production rather than from a decrease in outer membrane–peptidoglycan linkage (Wessel et al. 2013). An oprI mutant produced a 3-fold higher OMV level than the wild-type strain, while its PQS production was unaffected, indicating that the absence of OprI stimulated OMV biogenesis by reducing the outer membrane tethering to peptidoglycan. Finally, the OMV amount was unaffected in an oprL mutant compared to the wild-type strain (Wessel et al. 2013). The different mechanisms by which OprF and OprI impact OMV formation and the lack of effect of OprL indicate that, although these three proteins likely form ternary complexes maintaining the outer membrane integrity (Navare et al. 2015), they can also act individually. To the best of our knowledge, it remains unknown if variations in OprF levels in wild-type cells are sufficient to modulate OMV formation.
Several proteome studies identified OprF as a constituent of OMVs produced by P. aeruginosa PAO1 in various conditions: liquid cultures, biofilms, antibiotic treatment (Choi et al. 2011; Maredia et al. 2012; Toyofuku et al. 2012; Couto et al. 2015; Park et al. 2015). OprF was found to be the second most abundant protein in OMVs from liquid cultures (Choi et al. 2011) and was among the 30 most abundant proteins in OMVs from biofilms (Couto et al. 2015). Park et al. (2015) observed that some outer membrane proteins can be preferentially incorporated or omitted into OMVs, supporting the notion of a specific protein packaging during OMV biogenesis. OprF was over-represented in biofilm and planktonic OMVs at the 24-h time point (Park et al. 2015), suggesting that OprF could play important roles in OMV biogenesis. OMVs allow the transport of biological material from the parental bacterium to distal sites. In various Gram-negative bacteria, they were shown or proposed to display a variety of functions including transfer of antibiotic resistance, competition with other bacteria, stress response and bacterial survival, virulence factor delivery, bacterial adhesion and biofilm formation, nutrient and iron acquisition, cell communication, host cell invasion and modulation, and immune evasion (Kulp and Kuehn 2010; Kim et al. 2015; Orench-Rivera and Kuehn 2016; Pathirana and Kaparakis-Liaskos 2016). OprF could contribute to one or several of these functions, for example, given its involvement in the binding of P. aeruginosa to human cells (see above paragraph), it is tempting to hypothesize that OprF contributes to the interaction of OMVs with host cells. To our knowledge, the contribution of OprF or other individual proteins in OMV functions has not been reported yet.
OprF involvement in quorum-sensing response
Quorum sensing (QS) is a cell density-dependent mechanism characterized by the production of diffusible extracellular signal molecules, which trigger cellular responses, such as the production of virulence factors in the case of P. aeruginosa (Venturi 2006; Williams and Camara 2009; Papenfort and Bassler 2016). Pseudomonas aeruginosa produces two types of N-acyl-homoserine lactones, N-(3-oxododecanoyl)-L-homoserine lactone (3-oxo-C12- HSL), and N-butyryl-L-homoserine lactone (C4-HSL) (Venturi 2006) and two 4-hydroxy-2-alkylquinolines (HAQs), which includes 3,4-dihydroxy-2-heptylquinoline, also known as the Pseudomonas quinolone signal (PQS), and its precursor, 4-hydroxy-2-heptylquinoline (HHQ) (Diggle et al. 2007; Dubern and Diggle 2008). Another molecule, termed IQS, has been proposed as well as the product of the ambBCDE locus (Lee et al. 2013). However, more recent data show that IQS is in fact aeruginaldehyde, a by-product of the pyochelin siderophore biosynthetic pathway rather than the product of the ambABCDE locus as proposed by Lee et al. (Ye et al. 2014; Rojas Murcia et al. 2015). In the las system, the synthase LasI produces 3-oxo-C12 HSL and the LasR regulator binds the signal molecule, activating several target genes, while the rhl system involves RhlI, the C4-HSL synthase and the RhlR response regulator. These two systems are inter-connected and activate together the production of numerous virulence factors including elastase, staphylolytic protease, exotoxin A, rhamnolipids, pyocyanin, lectins and superoxide dismutases (Venturi 2006; Williams and Camara 2009). On the other hand, the HHQ and PQS signal molecules interact with the PqsR/MvfR regulator and together trigger the production of virulence factors, including the LecB lectin and the phenazine compound pyocyanin (Diggle et al. 2007). Recently, it was found that MvfR is a truly global regulator of QS system in P. aeruginosa, regulating both rhl and las systems as well as genes for the defense against oxidative stress, highlighting the central role of the HHQ-PQS/MvfR system in P. aeruginosa (Maura et al. 2016).
The production of several QS-controlled virulence factors (pyocyanin, elastase, LecA lectin, exotoxin A) is strongly reduced in an oprF mutant (Fito-Boncompte et al. 2011). Accordingly, the production of PQS by the oprF mutant was found to be decreased while the production of HHQ was considerably increased (Fito-Boncompte et al. 2011). The conversion of HHQ to PQS involves a hydroxylation reaction realized by the PqsH enzyme, the gene of which is under the control of the LasR (Diggle et al. 2007). In line with the accumulation of HHQ, and the decreased production of 3-oxo-C12-HSL in the oprF mutant, the activity of a pqsH-lacZ fusion was decreased in the oprF mutant (Fito-Boncompte et al. 2011). An impact of OprF on PQS production was also reported by Wessel et al. (2013). but it was opposite: the PQS level was increased in an oprF mutant. This discrepancy could result from the use of different parental strains (the PAO1-derivative H103 by Fito-Boncompte et al. and PA14 by Wessel et al.), different culture conditions and/or different PQS quantification methods (LC/MS by Fito-Boncompte et al. and TLC by Wessel et al.). In a P. aeruginosa PAO1 pqsA mutant unable to produce HHQ and PQS, the transcription of oprF and sigX, the sigma factor gene upstream of oprF (see below) was 4-fold higher than in a wild-type background, suggesting an involvement of these QS regulators on transcriptional control of sigX (Gicquel et al. 2013).
OprF involvement in the perception of environmental cues
Several reports strongly support a role of OprF as an outer membrane protein involved in the perception of environmental signals, including some produced by the infected host. The innate immune system complement cascade is a first line of defense against pathogens and it involves the conversion of C3 to C3a and C3b, which typically recognizes a component at the surface of the pathogen, activating the C5 convertase and the formation of C5b-9 membrane attack complex (Joiner, Fries and Frank 1987). OprF is probably a P. aeruginosa C3b receptor since an oprF-negative mutant showed reduced C3b binding compared to the wild type while expression of oprF in E. coli increased the deposition of C3b (Mishra et al. 2015). Wu et al. (2005) also demonstrated that among all tested cytokines, OprF binds interferon-gamma (IFN-γ), inducing expression by P. aeruginosa of the LecA lectin, a virulence factor under the control of the Rhl QS system. In line with the increased lecA expression, the same authors found that IFN-γ also induces the rhlI gene encoding the C4-HSL synthase (Wu et al. 2005).
During an acute phase of inflammation, the serum amyloid protein (SAA) is produced by hepatocytes and it was found that it tightly binds to Gram-negative bacteria, including E. coli, Salmonella enterica Thyphimurium, Shigella flexneri, Klebsiella pneumoniae, Vibrio cholerae and P. aeruginosa (Hari-Dass et al. 2005). In E. coli, OmpA is bound by SAA, while OprF is likely to be the target in P. aeruginosa since an oprF mutant is unable to bind SAA (Hari-Dass et al. 2005).
OprF importance in acute and chronic infections
In one study, mice were infected with P. aeruginosa PAO1, either to cause sepsis (acute infection) or chronic wound infection, and total gene expression was compared with cells grown in mid-logarithmic phase in a MOPS-succinate medium under laboratory conditions (Turner et al. 2014). Under both infection conditions, oprF expression was found to be downregulated, especially in the case of chronic wound infection where the expression was decreased by a factor 7.7 (Turner et al. 2014). This result seems to be counterintuitive given the importance of OprF for the expression of virulence factors (Fito-Boncompte et al. 2011), but one should take other factors into account than the degree of expression in order to evaluate the contribution of a given gene in fitness during infection processes (Turner et al. 2014). One way to address this issue is the use of massive random transposon insertions in a Tn-seq approach by comparing the number of independent transposon insertions in one given gene when bacteria are grown in laboratory conditions or in vivo in acute or chronic infections. In their study, Turner et al. (2014) found that the number of insertions in the oprF gene was quasi null in the case of bacteria recovered from acute infections and zero in the case of bacteria from chronic wound infection, indicative of a high fitness cost caused by oprF inactivation, supporting the importance of OprF in the infection process. Another type of infection caused by P. aeruginosa is the CF lung chronic colonization (Folkesson et al. 2012). In a recent study, it was revealed that transposon insertions in the oprF gene were almost absent when P. aeruginosa PAO1 was grown in CF sputum medium indicating an important fitness cost as well, while in the case of PA14, no insertion was found suggesting that oprF is an essential gene in this strain, at least in this growth condition (Turner et al. 2015). Interestingly, the same study showed that sigX is essential in both strains for growth in sputum medium (Turner et al. 2015). In non-CF bronchiectasis, an important T-cell response to OprF protein and an immune-dominant HLA-restricted T-cell epitope of OprF was detected (Quigley et al. 2015).
Genomic environment and regulation of oprF transcription
A first transcriptional study suggested that oprF was constitutively transcribed as a single gene from a σ70-type promoter (Duchene et al. 1988). However, subsequent work showed that although the oprF predominant mRNA is monocistronic, a larger sigX-oprF transcript could be detected (Brinkman et al. 1999). RT-PCR experiments furthermore indicated transcriptional linkages between the estX, menG, cmaX, cfrX, cmpX and sigX genes (PA1771 to PA1776 in P. aeruginosa PAO1) lying upstream of oprF (PA1777) (Fig. 2C), which suggested that oprF was the last of a seven-gene operon (Brinkman et al. 1999), which remains to be confirmed. Remarkably, this genomic structure, including ppsA upstream of estX, is fully conserved, not only among all of the sequenced P. aeruginosa genomes, but also among all of the sequenced genomes of members of the Pseudomonas genus (572 genomes; Winsor et al. 2016) (Table 1), suggesting both genetic and functional links between all of the genes and the corresponding proteins. The genes in this cluster are predicted to encode mainly proteins with putative functions: this is the case for the EstX esterase, the MenG ribonuclease E inhibitor, the CorA-like inner membrane Mg2+ transporter protein CmaX, the CfrX hypothetical protein of unknown function and the CmpX putative small mechanosensitive channel of the MscS family (Winsor et al. 2016). By contrast, SigX is an ECF sigma factor sharing 49% of similarity with SigW from Bacillus subtilis, which is known to regulate the membrane fluidity by changing the branched-to-linear fatty acid ratio (Kingston et al. 2011). SigX has recently been characterized and was found to be a master regulator since the deletion of its gene affects the expression of more than 300 genes, leading to changes in the production of virulence traits and resulting in increased formation of biofilms (Gicquel et al. 2013; Blanka et al. 2014; Bouffartigues et al. 2015; Schulz et al. 2015). SigX is involved in de novo fatty acids biosynthesis, thus regulating membrane fluidity (Boechat et al. 2013; Blanka et al. 2014) and outer membrane biogenesis (Duchesne et al. 2013). Since SigX is activated in response to factors causing cell wall stresses (low osmolarity, high sucrose concentration, absence of OprF, D-cycloserine treatment), this ECF sigma factor has been proposed to represent a new envelope stress response regulator (Bouffartigues et al. 2012; Duchesne et al.2013, 2014, 2015; Blanka et al. 2014). Remarkably, SigX was shown to be involved in oprF transcription in P. aeruginosa H103 (a derivative of the PAO1 strain): two intertwined promoters have been identified upstream of oprF, one of them being SigX-dependent while the other one depends on the σ70 primary sigma factor (Fig. 2C) (Duchene et al. 1988; Brinkman et al. 1999; Bouffartigues et al. 2012). In addition, the cmpX promoter region was immuno-precipitated by SigX using a ChIP-seq approach, suggesting a direct influence of SigX also on cmpX expression (Blanka et al. 2014). Taken together, these data support the hypothesis of functional relations between the members of this highly conserved genomic structure among P. aeruginosa genomes, which have now to be further deciphered.
While SigX is involved in oprF transcription in P. aeruginosa H103 (Brinkman et al. 1999; Bouffartigues et al. 2012; Gicquel et al. 2013), this has not been clearly established for strain PA14 where oprF transcription was not significantly affected by a sigX mutation although the oprF promoter region was successfully immunoprecipitated by SigX using a ChIP-seq approach (Blanka et al. 2014). In strain PA14, overexpression of sigX led to a biphasic growth curve where cells reached a first plateau, followed by a decline phase resulting in swelling and death of the cells before a recovering phase leading to a new plateau (Boechat et al. 2013). During the first growth phase, the production of OprF was unchanged, but a moderate overproduction was observed when cells recovered (Boechat et al. 2013). In addition, a search for promoters of the ECF sigma factor AlgU led to the identification of a third oprF promoter lying within the sigX open reading frame (Firoved, Boucher and Deretic 2002) (Fig. 2C). AlgU, a homolog of the E. coli ECF σE, was especially investigated because its hyperactivity (in most of the cases resulting from a mutation in the mucA gene encoding the anti-sigma) leads to overproduction of the exopolysaccharide alginate inducing the mucoid phenotype (Boucher et al. 1997). Firoved et al. showed by primer extension analysis that this AlgU promoter region was active in a strain lacking MucA, which specifically inhibits AlgU (Schurr et al. 1996), resulting in a 2.5-fold increase in oprF transcription. The possibility that oprF is part of the AlgU regulon suggests that oprF is also highly transcribed during lung colonization in CF patients where mucA mutants frequently arise. Accordingly, OprF is considered as a marker of P. aeruginosa in infected CF patients (Hassett et al. 2002; Eichner et al. 2014). The existence of this AlgU-dependent promoter region has however been the object of controversies since in another study the activities of transcriptional oprF-lacZ fusions were not significantly increased in a mucA mutant background (Malhotra et al. 2000). It was further shown that this AlgU promoter region only made a 10% marginal contribution to oprF expression when bacteria were grown in LB medium (Bouffartigues et al. 2012). Moreover, the global transcription of oprF remained quite similar in mucA and algU mutant backgrounds, in which AlgU was strongly active or absent, respectively (Bouffartigues et al. 2012). Taken together, these results suggest that net oprF transcription might remain similar in mucoid (hyperactive AlgU leading to overproduction of the alginate exopolysaccharide) and in non-mucoid strains (weakly or not active AlgU) where SigX-dependent expression is higher (Bouffartigues et al. 2012). Noticeably, the localization of this AlgU-dependent promoter within the sigX ORF may suggest that oprF transcription initiation from the AlgU-dependent promoter may result in reduced sigX expression.
More recently, a binding site for the AmpR regulator belonging LysR family was identified in the close vicinity of the SigX-dependent promoter region using ChIP-Seq assays (Balasubramanian et al. 2014). AmpR is a global transcriptional regulator: it regulates resistance to different classes of clinically relevant antibiotics, either positively by increasing resistance to β-lactams and aminoglycosides or negatively by decreasing resistance to quinolones. AmpR plays a key role in determining P. aeruginosa virulence and physiology by regulating expression of transcriptional and posttranscriptional regulators that feed into critical global networks, such as QS, Gac-Rsm and iron uptake (Balasubramanian, Kumari and Mathee 2015). Remarkably, AmpR is an integral part of the stress response system in P. aeruginosa, and plays a central role in the cell wall recycling (Balasubramanian et al. 2012). Overall, the discoveries of a complex promoter organization (Brinkman et al. 1999; Firoved, Boucher and Deretic 2002; Balasubramanian et al. 2012; Bouffartigues et al. 2012; Balasubramanian, Kumari and Mathee 2015) revealed that oprF transcription is highly regulated and under control of the ECF sigma factors SigX and AlgU, and of the master regulator AmpR, the activity of which has been related to cell wall stress responses (Wood, Leech and Ohman 2006; Balasubramanian et al. 2012; Balasubramanian, Kumari and Mathee 2015).
Transcriptome and proteome studies revealed that oprF is always transcribed, explaining why it was originally thought to be constitutively expressed, although its level of expression is subjected to variations. Table 2 gives an overview of the several proteomic and transcriptomic studies in which oprF expression was found to be affected by a given condition. As already mentioned, OprF is 40-fold overproduced in anaerobic biofilm condition (Hassett et al. 2002) and in a synthetic medium (ASM) that mimics the environmental conditions found in the infected lung of CF patients, and which induces the formation of microcolonies (Sriramulu et al. 2005). However, a strongly reduced production of microcolonies and expression of oprF was observed when the same medium was deprived of amino acids, which are known to be present in the CF lung (Sriramulu et al. 2005). In the same article, production of the OprF protein was also shown to be highly increased in ASM containing amino acids using 2D protein gels, in line with the increased oprF transcription level. These results contrast with the data of a study using ASM in the strain PA14 where a 6-fold decrease of oprF expression was observed in biofilms formed after 96 h under anaerobic conditions (Tata et al. 2016). The conditions used in the two contrasting studies are however quite different: Sriramulu et al. grew the PAO1 strain in 24 wells in ASM under gentle agitation for 16 h, while Tata et al. used the strain PA14 and grew the cells under anaerobic conditions in the presence of KNO3 and high concentrations of iron (100 μM) for 96 h.
Conditions leading to oprF expression variations.
| Conditions . | Strain . | Fold change . | References . |
|---|---|---|---|
| Transcriptomics | |||
| sigX mutant | PAO1 | –4 | Gicquel et al. (2013) |
| oprD mutant | PA14 | >10 | Skurnik et al. (2013) |
| Chronic/MOPS | PAO1 | –7.6 | Turner et al. (2014) |
| Biofilm/planktonic | PA14 | –6.3 | Tata et al. (2016) |
| Anaerobic/aerobic | PA14 | –5.6 | Babin et al. (2016) |
| TP359 cationic peptide | ATCC 39324 mucoid | –10 | Dosunmu et al. (2016) |
| TP359 cationic peptide | ATCC 39324 non-mucoid | –5 | Dosunmu et al. (2016) |
| Proteomics | |||
| Biofilm LB-nitrate + O2/biofilm LB + O2 | PAO1 | 4.9 | Yoon et al. (2002) |
| Biofilm LB-nitrate + O2/biofilm LB – O2 | PAO1 | 39 | Yoon et al. (2002) |
| TP359 cationic peptide | ATCC 39324 mucoid | –5 | Dosunmu et al. (2016) |
| TP359 cationic peptide | ATCC 39324 non-mucoid | –3 | Dosunmu et al. (2016) |
| ASM + amino acids | PAO1 | >10 | Sriramulu et al. (2005) |
| sigX mutant | PAO1 | –2 to –8* | Duchesne et al. (2013) |
| sigX overexpression | PA14 | 4.4 | Boechat et al. (2013) |
| Alkanes exposure | PseA (keratitis) | –** | Hemamalini and Khare (2014) |
| Conditions . | Strain . | Fold change . | References . |
|---|---|---|---|
| Transcriptomics | |||
| sigX mutant | PAO1 | –4 | Gicquel et al. (2013) |
| oprD mutant | PA14 | >10 | Skurnik et al. (2013) |
| Chronic/MOPS | PAO1 | –7.6 | Turner et al. (2014) |
| Biofilm/planktonic | PA14 | –6.3 | Tata et al. (2016) |
| Anaerobic/aerobic | PA14 | –5.6 | Babin et al. (2016) |
| TP359 cationic peptide | ATCC 39324 mucoid | –10 | Dosunmu et al. (2016) |
| TP359 cationic peptide | ATCC 39324 non-mucoid | –5 | Dosunmu et al. (2016) |
| Proteomics | |||
| Biofilm LB-nitrate + O2/biofilm LB + O2 | PAO1 | 4.9 | Yoon et al. (2002) |
| Biofilm LB-nitrate + O2/biofilm LB – O2 | PAO1 | 39 | Yoon et al. (2002) |
| TP359 cationic peptide | ATCC 39324 mucoid | –5 | Dosunmu et al. (2016) |
| TP359 cationic peptide | ATCC 39324 non-mucoid | –3 | Dosunmu et al. (2016) |
| ASM + amino acids | PAO1 | >10 | Sriramulu et al. (2005) |
| sigX mutant | PAO1 | –2 to –8* | Duchesne et al. (2013) |
| sigX overexpression | PA14 | 4.4 | Boechat et al. (2013) |
| Alkanes exposure | PseA (keratitis) | –** | Hemamalini and Khare (2014) |
Several isoforms were observed after 2DE.
Fold change has not been indicated.
Conditions leading to oprF expression variations.
| Conditions . | Strain . | Fold change . | References . |
|---|---|---|---|
| Transcriptomics | |||
| sigX mutant | PAO1 | –4 | Gicquel et al. (2013) |
| oprD mutant | PA14 | >10 | Skurnik et al. (2013) |
| Chronic/MOPS | PAO1 | –7.6 | Turner et al. (2014) |
| Biofilm/planktonic | PA14 | –6.3 | Tata et al. (2016) |
| Anaerobic/aerobic | PA14 | –5.6 | Babin et al. (2016) |
| TP359 cationic peptide | ATCC 39324 mucoid | –10 | Dosunmu et al. (2016) |
| TP359 cationic peptide | ATCC 39324 non-mucoid | –5 | Dosunmu et al. (2016) |
| Proteomics | |||
| Biofilm LB-nitrate + O2/biofilm LB + O2 | PAO1 | 4.9 | Yoon et al. (2002) |
| Biofilm LB-nitrate + O2/biofilm LB – O2 | PAO1 | 39 | Yoon et al. (2002) |
| TP359 cationic peptide | ATCC 39324 mucoid | –5 | Dosunmu et al. (2016) |
| TP359 cationic peptide | ATCC 39324 non-mucoid | –3 | Dosunmu et al. (2016) |
| ASM + amino acids | PAO1 | >10 | Sriramulu et al. (2005) |
| sigX mutant | PAO1 | –2 to –8* | Duchesne et al. (2013) |
| sigX overexpression | PA14 | 4.4 | Boechat et al. (2013) |
| Alkanes exposure | PseA (keratitis) | –** | Hemamalini and Khare (2014) |
| Conditions . | Strain . | Fold change . | References . |
|---|---|---|---|
| Transcriptomics | |||
| sigX mutant | PAO1 | –4 | Gicquel et al. (2013) |
| oprD mutant | PA14 | >10 | Skurnik et al. (2013) |
| Chronic/MOPS | PAO1 | –7.6 | Turner et al. (2014) |
| Biofilm/planktonic | PA14 | –6.3 | Tata et al. (2016) |
| Anaerobic/aerobic | PA14 | –5.6 | Babin et al. (2016) |
| TP359 cationic peptide | ATCC 39324 mucoid | –10 | Dosunmu et al. (2016) |
| TP359 cationic peptide | ATCC 39324 non-mucoid | –5 | Dosunmu et al. (2016) |
| Proteomics | |||
| Biofilm LB-nitrate + O2/biofilm LB + O2 | PAO1 | 4.9 | Yoon et al. (2002) |
| Biofilm LB-nitrate + O2/biofilm LB – O2 | PAO1 | 39 | Yoon et al. (2002) |
| TP359 cationic peptide | ATCC 39324 mucoid | –5 | Dosunmu et al. (2016) |
| TP359 cationic peptide | ATCC 39324 non-mucoid | –3 | Dosunmu et al. (2016) |
| ASM + amino acids | PAO1 | >10 | Sriramulu et al. (2005) |
| sigX mutant | PAO1 | –2 to –8* | Duchesne et al. (2013) |
| sigX overexpression | PA14 | 4.4 | Boechat et al. (2013) |
| Alkanes exposure | PseA (keratitis) | –** | Hemamalini and Khare (2014) |
Several isoforms were observed after 2DE.
Fold change has not been indicated.
Post-transcriptional regulation of oprF
In E. coli, the production of the OprF homolog OmpA is controlled at a post-transcriptional level by small non-coding RNAs (sncRNAs) MicA and RybB, binding to the Hfq RNA chaperone, ensuring a concerted expression of proteins involved in the control of outer membrane permeability (Johansen et al. 2006; Bossi and Figueroa-Bossi 2007; Udekwu and Wagner 2007). In E. coli, MicA is an antisense RNA of ompA mRNA, causing its destabilization in the presence of Hfq and the micA gene is under the control of RpoE ECF sigma, an AlgU homolog (Udekwu and Wagner 2007). In E. coli and Salmonella, the folding status of porins is monitored by the RpoE signaling system (Skovierova et al.2006; Bossi and Figueroa-Bossi 2007). Although we do not know at this stage whether a similar situation occurs in the post-transcriptional regulation of porin genes in P. aeruginosa, it is interesting to mention one report of a control of oprF mRNA translation via RsmA/RsmE in P. fluorescens CHAO (Crespo and Valverde 2009). It is therefore tempting to speculate for such a translational regulation of the oprF expression, in addition to the regulation of its transcription. However, one search for sRNAs in intergenic regions in the PAO1 genome failed to give any evidence of such small RNAs in the intergenic region between sigX and oprF (Gomez-Lozano et al. 2012). In a 2D-gel proteomic study comparing a wild-type P. aeruginosa PAO1 and its sigX mutant, it was found that the levels of some isoforms of OprF were decreased in the sigX mutant (Duchesne et al. 2013), suggesting also the existence of some post-translational modifications of the porin. Noticeably, the L-fucose-specific lectin LecB was shown to recognize OprF (Funken et al. 2012). In addition, considering that OprF is a sensor for human interferon gamma (IFNγ) (Wu et al. 2005), and that a fucosyl-residue is required for recognition of IFNγ by the receptor (Lundell and Narula 1994), it is possible that the fucose-specific LecB may act as an adaptor to mediate recognition of this cytokine by OprF on the bacterial surface (Funken et al. 2012). These data further suggest that OprF could be glycosylated, a hypothesis that is further strengthened by the recent assumption that OprF is a sialoglycoprotein having both 2,6- and 2,3-linkages (Khatua et al. 2014).
THE SMALL OprG PORIN
OprG with only eight β-strands is the second smallest Pseudomonas aeruginosa porin with a molecular mass of 25.2 kDa (Gensberg et al. 1999; McPhee et al. 2009; Touw, Patel and Van den Berg 2010). Its structure, presented in Fig. S1, shows the presence of large loops extending to the exterior of the outer membrane. OprG belongs to the OmpW family, which includes the Escherichia coli OmpW (Hong et al. 2006), Caulobacter crescentus OmpW (Benz et al. 2015) and the closest homolog OmpW from Pseudomonas putida (Walzer, Rosenberg and Ron 2009). OprG has been suggested to allow the transport of hydrophobic molecules via a lateral opening in the barrel wall (Touw, Patel and Van den Berg 2010). The OprG channel is however too small to allow the diffusion of small molecules like amino acids, despite that transport of alanine, glycine, serine and valine has been determined in liposome swelling assays (Kucharska et al. 2015). Although the oprG mRNA is monocistronic, two of the upstream genes are involved in transport of amino acids and peptides (Kucharska et al. 2015). Another possible and likely function of OprG is the diffusion of Fe2+ under microaerobic conditions where this form of iron is predominant (Yates, Morris and Brown 1989). A strong support for the hypothesis that OprG might be involved in the diffusion of Fe2+ comes from the recent report on the involvement of Acinetobacter baumanii OmpW, an OprG homolog, in the transport of ferrous iron (Catel-Ferreira et al. 2016). Using an oprG::xylE fusion, McPhee et al. (2009) showed that the oprG gene transcription is induced by high iron levels under anaerobic conditions and that the Anr regulator is involved in this regulation. The presence of an Anr box 71 nucleotides upstream of oprG ATG was later confirmed (Trunk et al. 2010). The same authors confirmed a strong upregulation of oprG (20 x) under anaerobic growth conditions. More recently, using artificial medium mimicking CF lung conditions, i.e. low oxygen tension and high cell density, but in absence of added nitrate, other authors showed a strong decrease of oprG expression in a Δanr mutant compared to wild type in strains PAO1 (–18 x) and J215 (–35 x), confirming again the involvement of Anr in the regulation of oprG transcription (Hammond et al. 2015).
Zaborin et al. (2012) demonstrated the induction by opioids of virulence factors production by P. aeruginosa and one of the genes which was found to be expressed at higher level is oprG. In a RNA-seq study, Dötsch et al. (2012) found that the level of transcription of oprG is reduced by almost a factor 5 in 48-h biofilms of P. aeruginosa PA14 grown in LB medium compared to 4-h planktonic cells. In another study, Chua et al. (2014) obtained a different result since they found that oprG expression is higher (by a factor 3) in biofilms compared to planktonic cells. This discrepancy might originate from the different conditions used to grow biofilms, LB medium in the Dötsch et al. study and a minimal medium plus glucose by Chua et al. An interesting aspect of the last study is the effect of cells dispersal from biofilms, which revealed that biofilm dispersed cells are in a different physiological state compared to planktonic cells, characterized by a decreased production of the siderophore pyoverdine and an increased virulence (Chua et al. 2014). Treatment of planktonic cells with sodium nitroprusside (SNP), which is normally used to disperse biofilms, caused a decrease of expression of oprG by a factor 12 (Chua et al. 2014). Exposure to steady-state levels of H2O2 causes a decrease in oprG transcription by a factor 8 while no changes in the expression of other porin genes could be observed (Deng et al. 2014). In one study, exposure of P. aeruginosa cells to respiratory epithelial cells caused a decrease in the expression of oprG by a factor 4 (Chugani and Greenberg 2007). Finally, a transcriptome comparison of a spontaneous small colony variant (SCV) of P. aeruginosa PAO1 with the original wild type revealed a 3-fold decrease of oprG transcription in early stationary phase of growth (Wei et al. 2011). The ECF sigma factor SigX, already known as a regulator of oprF, has also been found to be involved in the regulation of oprG, via ChIP-seq, RNA-seq and supported by the presence of a conserved SigX binding motif (Schulz et al. 2015). Interestingly, a conserved binding motif for the FecI ECF sigma factor, involved in the regulation of uptake of ferric citrate (Potvin, Sanschagrin and Levesque 2008; Cornelis, Matthijs and Van Oeffelen 2009), has also been confirmed by ChIP-seq analysis, in further support of the suggestion of OprG involvement in iron homeostasis (Schulz et al. 2015) (Table 1).
Like six other porins (OprE/OccK8, OprD/OccD1, OprB, OprF, OprQ/OccD6 and OprH), OprG has been identified as a component of OMVs produced by P. aeruginosa PAO1 in liquid cultures or in biofilms in three distinct proteome studies (Choi et al. 2011; Toyofuku et al. 2012; Couto et al. 2015). However, OprG is the only porin which has been identified as being under-represented in biofilm OMVs compared to outer membranes of the corresponding biofilm bacteria at two of the three studied time points (24 and 96 h, not at 48 h) (Park et al. 2015), but the significance of this observation remains unknown.
OprH, THE LOW-MAGNESIUM INDUCED PORIN AND ITS INVOLVEMENT IN ANTIBIOTIC RESISTANCE
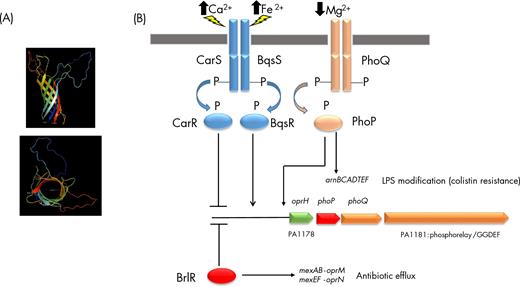
OprH is the smallest of the Pseudomonas aeruginosa porins, with a mass of only 21.6 kDa. Similar to OprG, OprH has eight β-strands and its interaction with LPS has been demonstrated (Edrington et al. 2011; Kucharska et al. 2016) (Fig. 3A, Fig. S1). The oprH gene (PA1178) forms an operon with phoP-phoQ and PA1181 (Fig. 3B). In P. aeruginosa, the PhoP-PhoQ and the PmrA-PmrB two-component regulatory systems are known to be induced upon Mg2+ starvation and PhoP-PhoQ to directly upregulate the production of OprH, resulting in increased resistance to the polycationic antibiotic polymyxin B (Macfarlane et al. 1999; Macfarlane, Kwasnicka and Hancock 2000; McPhee et al. 2006; Wei et al.2011; Olaitan, Morand and Rolain 2014). Polyamines have also been described to induce both the PhoP-PhoQ two-component system, the OprH porin and the PA3552-PA3559 LPS modification genes (arn genes), resulting in higher resistance to polymyxin and aminoglycosides (Kwon and Lu 2006). In agreement with this, addition of the polyamines agmatine and putrescine increases the level of oprH transcription by a factor of 9 and 7, respectively (Chou et al. 2008). Interestingly, the presence of high concentrations of Ca2+ ions has a strong negative effect (factor -58) on phoP-phoQ and hence on oprH expression (Guragain et al. 2016). The same authors found that calcium homeostasis is regulated by the CarSR two-component system and a phylogenetic analysis revealed that the CarR response regulator is closely related to PhoP, suggesting a possible cross-talk between magnesium and calcium homeostasis regulation (Guragain et al. 2016) (Fig. 3B). One possible explanation is that OprH stabilizes the outer membrane by interacting with LPS and that high levels of Ca2+ (or Mg2+) could substitute for OprH by directly interacting with LPS, strengthening the outer membrane, hence causing a decreased abundance of OprH. In line with this hypothesis, Kreamer et al. identified a two-component system, BqsSR in P. aeruginosa, which responds to Fe2+ and promotes the expression of genes for cation tolerance, including genes for polyamine synthesis and uptake (Kreamer et al.2012; Kreamer, Costa and Newman 2015). In their RNA-seq analysis, they demonstrated that the phoPQ operon including oprH is under the control of the BqsSR two-component system (Kreamer, Costa and Newman 2015). When reading the publication of Guragain et al., we were surprised to realize that CarSR and BqsSR are encoded by the same genes (PA2656 and PA2657 in strain PAO1), which indicates that the same two-component system responds to both calcium and ferrous iron, but in an inverse relationship. It is indeed puzzling that Fe2+ shock results in an increased oprH expression (Kreamer, Costa and Newman 2015) while high concentrations of Ca2+ cause a decrease in the level of transcription of the oprH gene (Guragain et al. 2016). One possible explanation is that the two cations cause different degrees of phosphorylation of the sensor and the response regulator, a hypothesis worth investigating in the future.
OprH structure scheme and regulation of oprH transcription. (A) Schematic representation (PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC) viewed from the side (top) and from the extracellular environment (bottom) of OprH based on a 3D model predicted by I-TASSER (Yang and Zhang 2015; Yang et al. 2015). (B) Low level of Mg2+ is perceived by the TCS PhoP/PhoQ, in which the sensor PhoQ is submitted to autophosphorylation, leading to phosphorylation of its cognate regulator PhoP. The operon oprH/phoP/phoQ/PA1181 is subjected to a positive feedback since the oprH promoter region is a direct target of PhoP. In addition, perception of high levels of Ca2+ or Fe2+ by CarS/BqsS, respectively, converge on CarR/BqsR, which acts as a negative and positive regulator, respectively, on oprH transcription. Finally, the c-di-GMP responsive transcription factor BrlR is a negative regulator of the oprH operon. Taken together, these data suggest that regulation of oprH-phoP-phoQ-PA1181 is closely related to antibiotic resistance, since arnBCADTEF that is involved in LPS modification leading to colistin resistance is positively regulated by PhoP, and BrlR is a positive transcriptionnal regulator of the two major efflux pumps MexAB-OprM and MexEF-OprN. In addition, PA1181 is a predicted c-di-GMP cyclase, suggesting a link between oprH-phoP-phoQ-PA1181 and biofilm formation. Remarkably, PA1181 was shown to belong to the SigX regulon (Blanka et al. 2014), a newly proposed cell wall stress responsive sigma factor.
In a study on a spontaneous gentamycin-resistant SCV of P. aeruginosa strain PAO1, Wei et al. (2011) found larger amounts of the OprH protein in the SCV compared to the wild type, in agreement with the transcriptomic data showing an increased expression of the oprH gene in both early (4-fold) and late stationary phase of growth (5-fold). This effect was probably the result of a 39-bp deletion in the phoQ sensor open reading frame, which could explain the higher expression of PhoP-dependent genes, including oprH (Wei et al. 2011).
BrlR, a c-di-GMP-binding MerR-like regulator, which is known to be an activator of antibiotic efflux pumps (Liao and Sauer 2012; Liao, Schurr and Sauer 2013; Chambers et al. 2014), was found to repress the expression of the oprH-phoP-phoQ-PA1181 operon by binding directly upstream of the oprH gene, resulting in increased sensitivity to colistin (Chambers and Sauer 2013). When P. aeruginosa is in contact with bronchial epithelial cells, expression of the oprH-phoP-phoQ operon is strongly enhanced, by a factor more than 5000 in one study (Gellatly et al. 2012) whereas an earlier report by Chugani and Greenberg (2007) showed a decreased expression of oprH upon contact with lung epithelial cells. This apparent contradiction could be explained by the different approaches used in the two studies: while Chugani and Greenberg looked at the global gene expression upon contact with epithelial cells, Gellatly et al. specifically looked at the expression of epithelium adherent cells. To add to the complexity of oprH expression regulation, the RpoS sigma factor was found by ChIP-seq analysis to bind the upstream intergenic region, which was confirmed by the presence of a conserved RpoS binding motif (Schulz et al. 2015) (Table 1).
Recently, it has been demonstrated that OprH binds the surfactant protein A (SP-A), a lung lectin of the collectin family involved in innate immunity by promoting the bacteria clearance via phagocytosis by alveolar macrophages (Qadi et al. 2016). Direct binding of SP-A to OprH was also confirmed by fluorescent ligand binding (Qadi et al. 2016).
THE GLUCOSE-SPECIFIC PORIN GENES oprB AND oprB2
The oprB gene encodes a glucose-specific porin with 18 β-sheets (Wylie and Worobec 1994, 1995) (Fig. 4A) and is in an operon with four other genes, forming the gltBFGKoprB operon as predicted in the Pseudomonas.com database (Winsor et al. 2016) by the DOOR prokaryotic operon prediction tool (Mao et al. 2009) (Fig. 4B). GltB is predicted to be a periplasmic binding protein (Lewenza et al. 2005) and the gltFGK genes encode the components of an ABC transporter, GltF and GltG forming a sugar permease while GltK is the ATP binding subunit of the transporter. Although not predicted to be in the same operon, the upstream genes are probably involved in the regulation of glucose uptake as well. The gtrS gene is separated by a large intergenic region (523 nucleotides) from gltB and it encodes together with the neighboring gene gltR a glucose-sensitive two-component system in which GtrS and GltR are the histidine kinase and the response regulator, respectively (Sage, Proctor and Phibbs 1996; O’Callaghan et al. 2012; Daddaoua et al. 2014). The first gene of this second operon is edd encoding the phosphogluconate dehydratase, forming the edd-glK-gltR-gtrS predicted operon (Mao et al. 2009). Upstream of the edd gene, and in the opposite direction, the gapA gene leads to a monocistronic mRNA and encodes the glyceraldehyde triphosphate dehydrogenase. The response regulator GltR acts at three promoters, upstream of the edd, the glk and the oprB genes (Fig. 4B). The GtrS protein is more abundant when the cells have been exposed to epithelial cells and inactivation of the gtrS gene leads to a reduced cytotoxicity towards airway epithelial cell line, in line with the finding that the GtrS-GltR two-component system is involved in the induction of type III secretion system in Pseudomonas aeruginosa (O’Callaghan et al. 2012). It was also determined that this effect on type III secretion was partly dependent on OprB, linking oprB expression and virulence in P. aeruginosa since GltR also regulates the toxA gene for exotoxin A production (O’Callaghan et al. 2012; Daddaoua et al. 2014). It is also worth to mention that exposure to human respiratory epithelial cells results in increased expression of oprB by a factor of almost 4 (Chugani and Greenberg 2007). Another regulator, Anr, which regulates the shift from aerobiosis to anaerobiosis, also binds to a region upstream of the gltB gene, positively regulating its expression (Trunk et al. 2010).
OprB structure and regulation of oprB transcription. (A) Schematic representation (PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC) viewed from the side (left) and from the extracellular environment (right) of OprB based on a 3D model predicted by I-TASSER (Yang and Zhang 2015; Yang et al. 2015). (B) Sensing of 2 ketogluconate (2KG) by the histidine kinase GtrS results in phosphorylation of the GltR transcriptional regulator, which in turn negatively affects transcription of oprB, PA3190, glk and gapA, these genes lying in the close vicinity of the TCS encoding genes. In addition, GltR is also a negative regulator of toxA expression, suggesting a relation between glucose metabolism and virulence in P. aeruginosa.
The expression of oprB was found to be strongly downregulated (more than 50 times) in 96-h biofilms compared to planktonic cells under anoxic conditions (Tata et al. 2016). In a proteomic analysis, Duchesne et al. (2013) found that the SigX ECF sigma factor has also a positive effect on the level of the OprB porin production. The expression of oprB has also been found to be downregulated in the presence of ASM (Fung et al. 2010). In their survey of sigma factors regulons (sigmulons), Schulz et al. (2015) found one RpoS-binding motif upstream of oprB, supported by the results of their RNA-seq analysis (Table 1). In the genome of P. aeruginosa PAO1, there is a second gene, oprB2, coding for a protein of same size, which is 95% identical to OprB. This gene is predicted to be in an operon with the gcd gene encoding the glucose dehydrogenase and a gene (PA2289) encoding a TonB-dependent receptor. Interestingly, this TonB-dependent outer membrane receptor is not predicted to be regulated by an ECF sigma factor or by Fur, suggesting that it is not involved in iron uptake (Cornelis, Matthijs and Van Oeffelen 2009). The function of oprB2 and its neighboring genes is presently unknown.
THE OprO AND OprP PHOSPHATE-SPECIFIC PORINS
Both porin genes are transcribed as monocistronic mRNAs and follow each other (oprP: PA3279, oprO: PA3280) (Winsor et al. 2016). The two proteins share a high degree of similarity (76% identity and 16 conservative substitutions) (Siehnel, Egli and Hancock 1992) and are immunologically cross-reactive (Hancock et al. 1992). Their structures are also highly similar and the crystal structure of OprP has been solved at a 1.9 Å resolution (Moraes et al. 2007) (Fig. 5A, Fig. S1). Despite this high level of similarity, even at structural level, the two porins display different specificities, since OprP shows a preference for Pi monophosphate while OprO is more specific for di- and poly-phosphates (Hancock et al. 1992). The protein has an arginine ladder extending from the extracellular surface down to a constriction zone where Pi is coordinated while lysine residues coat the inner periplasmic surface (Moraes et al. 2007; Pongprayoon et al. 2009). There are two adjacent binding sites for Pi, W1 and W2, in OprP and the higher selectivity for phosphate compared to chloride is conferred by the lysine residues (Pongprayoon et al. 2009). Interestingly, there is as well an aspartate residue (D94) in the binding site, which seems counterintuitive, but mutation to a positively charged residue leads to a higher affinity binding of phosphate, but causes a decreased release of the ligand; the role of this aspartate residue is therefore proposed to decrease the residence time of Pi (Modi et al. 2015).
OprO, OprP and OpdH (OccD5), structure and regulation. (A) Schematic representation (PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC) viewed from the side (left) and from the extracellular environment (right) of OpdH and OprP based on their X-ray crystal structure (PDB 3T20 and 2O4V, respectively), and of OprO based on a 3D model predicted by I-TASSER (Yang and Zhang 2015; Yang et al. 2015). (B) PhoB is a positive regulator of oprO and oprP expression. By contrast, TctD is a repressor of oprO and oprP, as well as of its own expression and that of opdH (occD5). In presence of the OpdH (OccD5) substrate, cis-aconitate, the repression exerted by TcdT is alleviated, leading to transcription of opdH (occD5)-tctA-tctB-tctC, tctD-tctE operons, oprO and oprP.
As already mentioned, OprO is a channel for PPi while OprP binds preferentially Pi. The difference is to be found by the presence in the central constriction of two tyrosine residues in OprP (Y62 and Y114) while in OprO there is a phenylalanine at position 62 and an aspartate at position 114. Interestingly, permutations of these amino acids between the two phosphate porins resulted in changed specificities (Modi et al. 2015). Both oprO and oprP genes are upregulated when phosphate is limiting and upon binding of the PhoB regulator to pho boxes upstream of the two porin genes (Siehnel, Worobec and Hancock 1988; Siehnel, Egli and Hancock 1992). Reciprocally, oprO and oprP are downregulated in a mutant deleted of phoB compared to wild type (Bielecki et al. 2015). Another regulator, TctD, was found to control genes belonging to the PhoB regulon, including oprO, which is positively regulated by PhoB, but negatively by TctD (Bielecki et al. 2015). Interestingly, TctD is a regulator of the opdHtctCBA (occK5) operon where it acts as a repressor in the absence of the specific porin substrate (Tamber et al. 2007; Bielecki et al. 2015) (Fig. 5B).
Both oprP and oprO are upregulated when P. aeruginosa is in contact with lung epithelial cells, although this effect is even more pronounced in the case of oprO (by a factor 143 versus 10 for oprP) (Chugani and Greenberg 2007). In a study comparing the gene expression of P. aeruginosa PA14 cells in a healthy volunteer blood or in burn patients blood, it was found that in the last condition both oprP and oprO are downregulated (–8 x and –50 x, respectively) as well as the components of the PhoB regulon (Kruczek et al. 2016), suggesting that this downregulation is PhoB dependent. In line with this, both genes’ expression is downregulated in a mutant deleted of phoB compared to wild type (Bielecki et al. 2015). The RpoH sigma factor involvement in the regulation of oprP (and probably oprO) has been detected by ChIP-seq and the presence of an RpoH-binding motif has been confirmed (Schulz et al. 2015) (Table 1).
THE OprD (OccD1) BASIC AMINO ACID PORIN
OprD (OccD1) represents the second major porin protein in the outer membrane of Pseudomonas aeruginosa (Fig. 6A) and is also one of the most investigated because of its involvement in the entry of carbapenem antibiotics, especially imipenem and meropenem (Trias and Nikaido 1990a,b; Quinn et al. 1991; Yoneyama, Yamano and Nakae 1995). That OprD (OccD1) serves as the gate through which imipenem enters the P. aeruginosa cell was suspected by the analysis of imipenem-resistant mutants which did not produce detectable amount of the porin as observed by western blot analysis (Yoneyama and Nakae 1993; Wolter, Hanson and Lister 2004).
Regulation of oprD (occD1) and opdT (occD4). (A) Schematic representation (PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC) viewed from the side (left) and from the extracellular environment (right) of OprD (OccD1) based on its X-ray crystal structure (PDB 4FOZ), and of OpdT (OccD4) based on a 3D model predicted by I-TASSER (Yang and Zhang 2015; Yang et al. 2015). (B) Expression of oprD (occD1) is activated via the arginine responsive regulator ArgR and repressed directly by the zinc responsive CzcR. Repression of oprD (occD1) transcription by the copper-sensitive regulator CopR and by MexT, the activator of the mexEF-oprN efflux pump encoding genes, is suspected. Furthermore, regulation occurs as well at a post-transcriptional level involving the small RNA CrcZ and the Hfq RNA chaperone (in blue).
The first identified real physiological function of OprD (OccD1) was the transport of basic amino acids (arginine, lysine, histidine, ornithine) and peptides such as Ala-Lys, Lys-Ala, Thr-Ser-Lys or Pro-Phe-Gly-Lys, but not Thr-Lys-Pro, suggesting that peptides containing an internal charged amino acid are not taken up (Trias and Nikaido 1990a). It has also been suggested that OprD (OccD1) could be involved in the uptake of gluconate, but not glucose (Huang and Hancock 1993). The porin, reconstituted in planar lipid bilayers, bound imipenem via its second external loop (Huang and Hancock 1996). Mutations resulting in non-expression of oprD (occD1) result in imipenem resistance, but other mutations involving amino acid substitutions without affecting transcription also cause carbapenem resistance (Richardot et al. 2015). Recently, it was found that an in-frame deletion within the oprD (occD1) gene led to expression of a smaller protein, which resulted in carbapenem resistance while the capacity of transporting arginine was maintained (Kos, McLaughlin and Gardner 2016), although this could be due to the involvement of a second porin (OpdP/OccD3) in the uptake of arginine (see below). Although both meropenem and imipenem can be bound by the porin, mutants devoid of oprD (occD1) were only resistant to imipenem, but had an unchanged susceptibility to meropenem, suggesting the existence of an alternative uptake route for this last antibiotic (Perez et al. 1996).
OprD (OccD1) was the protein showing the greatest over-representation in OMVs from biofilm samples compared to the outer membrane of bacteria at all three investigated time points (Park et al. 2015). This over-representation was specific of the biofilm lifestyle since OprD (OccD1) was not specifically over-represented in planktonic OMVs. The authors proposed that packaging OprD (OccD1) into biofilm OMVs might be a way for P. aeruginosa to reduce the porin levels in its outer membrane, thereby lowering the bacterial sensitivity to carbapenem (Park et al. 2015). In Escherichia coli, OMVs were shown to protect bacteria from polymyxin B and colistin by adsorbing these antimicrobial peptides (Manning and Kuehn 2011). One could similarly hypothesize that high OprD (OccD1) levels in OMVs deplete OprD in the outer membrane, allowing carbapenem accumulation into OMVs, and lowering the concentration of free carbapenem in bacterial cells.
Purified OprD (OccD1) also displayed a protease activity while an amino acid triad characteristic of serine proteases (His, Asp, Ser) was identified and mutants constructed by site directed mutagenesis confirmed the protease activity of the porin (Yoshihara et al.1996, 1998). The structure of OprD (OccD1) has been determined, and, like in other members of the OccD/OprD family, it has 18 β-sheets and a constricted narrow pore containing a positively charged amino acids ladder on one side and another region of electronegative charged pocket on the other side (Biswas et al. 2007). The production of OprD (OccD1) is induced by arginine, glutamate or alanine as sole carbon sources when the cells are grown in minimal medium (Ochs et al. 1999b). The presence of the preferred carbon source succinate suppressed this induction, suggesting the involvement of a catabolite suppression mechanism (Ochs et al. 1999a).
The AraC family ArgR regulator was found by electrophoretic mobility shift assay to bind to the operator sequence of oprD (occD1) and ArgR protected a 47-bp region from DNAse I digestion, which included a consensus binding site for the regulator (Ochs et al. 1999a) (Fig. 6B, Table 1). Although the induction of oprD (occD1) by arginine was lost in an argR mutant, the glutamate-induced induction was still observed suggesting an alternative regulatory mechanism (Ochs et al. 1999a). MexT, a LysR family regulator, is an activator of the mexEF-oprN efflux pump and its overexpression has an inverse effect on oprD (occD1) expression, a phenomenon first observed in nfxC mutants, which were further identified as being mutants overexpressing the efflux pump (Kohler et al. 1999a,b) (Fig. 6B). Exposure to copper or zinc was found to cause a decrease in oprD (occD1) expression and led to undetectable levels of OprD (OccD1) protein (Caille, Rossier and Perron 2007). In 96-h biofilms grown under anoxic conditions, a strong decrease of oprD (occD1) expression was observed, which could partly explain the higher resistance to meropenems of biofilms (Tata et al. 2016). A similar effect, i.e. decreased expression of oprD (occD1), was observed in 48-h biofilms compared to planktonic cells (Dötsch et al. 2012). In line with this observation, a strong decrease in oprD (occD1) expression is observed in chronic wounds compared to MOPS-succinate grown cells (Turner et al. 2015).
The P. aeruginosa two-component CzcSR system is induced after exposure to metals and is involved in the resistance against copper and zinc via the induction of the CzcCBA metal efflux pump (Caille, Rossier and Perron 2007). Overproduction of CzcR after exposure to zinc and of another regulator involved in copper resistance, CopR, resulted in strongly decreased production of OprD (OccD1), the effect being stronger at post-transcriptional compared to transcriptional level (Caille, Rossier and Perron 2007). Using chromosome immunoprecipitation experiments (ChIP), it was demonstrated that CzcR directly binds in the control region of oprD, clearly establishing CzcR as a negative regulator of oprD (Dieppois et al. 2012) (Fig. 6B, Table 1). More recently, it has been shown that the Hfq RNA chaperone is required for binding of the CzcR regulator to the operator region of the porin gene (Ducret et al. 2016). Hfq binds preferentially to the CrcZ small RNA in P. aeruginosa PA14 and deletion of hfq leads to overexpression of oprD (occD1) as well as oprQ (occD6), suggesting a dual level of regulation, transcriptional via CzcR and post-transcriptional via the small RNA CrcZ and Hfq (Ducret et al. 2016; Pusic et al. 2016) (Fig. 6B, Table 1).
Twenty-six 2,3- and 2,6-linked sialoglycoproteins have been detected in P. aeruginosa exposed to human serum, and OprD (OccD1) is one of them, the others being OprG, OprF and OprQ (OccD6) (Khatua et al.2014). Interestingly, OprD (OccD1) sialylation has been found to impair its interaction with β-lactam antibiotics (Khatua et al. 2014). Analysis of 55 clinical and environmental P. aeruginosa isolates oprD (occD1) sequences revealed a mosaic structure of OprD (OccD1) where stretches of sequences have apparently been exchanged, and, in one isolate, an important sequence variability typical of a hypermutator phenotype has been observed (Pirnay et al. 2002).
Surprisingly, using a high-throughput Tn-seq analysis of P. aeruginosa PA14 strains, a vast majority of mutants with higher dissemination in a mouse model of infection were observed to lack oprD (occD1) expression, which not only did result in carbapenem resistance, but also in increased in vivo fitness (Skurnik et al. 2013). Strains lacking oprD (occD1) expression also showed higher resistance to low pH or normal human serum exposure while having an increased cytotoxicity towards human macrophages (Skurnik et al. 2013). This increased fitness is remarkably only observed for insertions in oprD (occD1) since all other Tn insertions in all the other outer membrane protein genes resulted in decreased rather than increased in vivo fitness (Skurnik et al. 2013). A RNA-seq analysis of PA14 wild type and Tn-oprD mutant did not reveal differences in expressed genes in vitro, while in vivo 97 genes were found to be silent in the mutant and, conversely, 60 genes had transcription levels increased more than 10-fold in the mutant (Skurnik et al. 2013). Interestingly, the oprF, oprG and oprQ (occD6) genes were more than 10 times more expressed in the oprD (occD1) mutant strain in the in vivo situation (Skurnik et al. 2013). In this context, it is tempting to link this observation with the fact that OprF is needed for full virulence of P. aeruginosa (Fito-Boncompte et al. 2011), which could partly explain why the oprD (occD1) mutant is more invasive and virulent. Similarly to OprF, whose production is lowered in a sigX mutant, the levels of the OprD (OccD1) protein were also found to be strongly decreased in a proteomic analysis comparing the ΔsigX and WT P. aeruginosa PAO1 (Duchesne et al. 2013). The same decrease in oprD (occD1) transcription was observed in a transcriptome analysis comparing the sigX mutant versus the wild type (Gicquel et al. 2013). The involvement of SigX in oprD (occD1) transcription has been further suggested by ChIP-seq and a motif was detected in the oprD (occD1) promoter region (Schulz et al. 2015) (Fig. 7). Finally, the involvement of RpoN sigma factor in the regulation of oprD (occD1) expression has been determined by ChIP-seq and RNA-seq analysis although no characteristic motif could be detected (Schulz et al. 2015) (Table 1, Fig. 7).
Regulation of porin genes by sigma factors. The involvement of FecI, FecI2, AlgU, RpoH, RpoS, SigX, FliA and RpoN sigma factors in regulation of porin genes is indicated by the corresponding colored lines, according to ChIP data (Schulz et al. 2015).
THE OTHER MEMBERS OF THE OprD/Occ FAMILY OF PORINS
The proteins of the members of the OprD (Occ) family share a high degree of similarity (46%–57%) (Fig. 8), but they show different substrate specificities (Tamber and Hancock 2006; Tamber, Ochs and Hancock 2006). In one study, eight porin genes belonging to the OprD (Occ) family were tested for their substrate specificity using nine different compounds (glucose, cis-aconitate, arginine, glycine-glutamate, pyroglutamate, tyrosine, vanillate, proline and histidine) (Tamber, Ochs and Hancock 2006). The three porin genes corresponding to the OpdK (OccK) subfamily (Fig. 8), opdH (occK5), opdK (occK1) and opdO (occK3), were specifically induced by their respective substrates (cis-aconitate and vanillate for opdH (occK5), vanillate for opdK (occK1) and pyroglutamate for opdO (occK3) (Tamber, Ochs and Hancock 2006)). Conversely, the five genes corresponding to members of the OprD/OccD subfamily, oprD (occD1), opdP (occD3), opdC (occD2), opdB (occD7) and opdT (occD4), showed a constitutive expression in the presence of all the tested substrates, with a slightly higher induction of opdP (occD3) in the presence of arginine (Tamber and Hancock 2006; Tamber, Ochs and Hancock 2006). Interestingly, OpdP (OccD3) shows the highest percentage of similarity with OprD (OccD1) (51%), suggesting that it could compensate for the absence of OprD (OccD1) for the uptake of arginine. This was confirmed by the observation that a double oprD/opdP (occD1/occD3) mutant grew less well in the presence of arginine as a carbon source compared to single mutants or to the wild type, which was further confirmed by a reduced uptake of radiolabeled arginine by the double mutant (Tamber and Hancock 2006). These results suggest that (i) the members of the OpdK (OccK) subfamily may have evolved a stricter substrate specificity, despite the presence of a larger pore in the members of this family (Eren et al.2012, 2013), and (ii) there is some substrate utilization redundancy between the porins of the OccD subfamily, similar to what is observed among the members of the TonB-dependent siderophore receptors (Ghysels et al.2004, 2005). Further characterization of OpdH (OccK5), the porin which is induced by cis-aconitate, confirmed the gene expression induction by cis-aconitate, but also by isocitrate and citrate (Tamber et al. 2007). The opdH (occK5) gene is the first of an operon with tctCBA where TctC is a periplasmic binding protein and the TctB and TctA proteins are the inner membrane components of a tri-carboxylate transporter (Fig. 5B). Transcribed in the opposite orientation are the two genes, tctDE, corresponding to a two-component system. Inactivation of tctD encoding the response regulator caused a derepression of the opdH (occK5) operon in the absence of inducer, although it did not result in the entire suppression of the induction by the substrate of OpdH (OccK5), suggesting another level of regulation (Tamber et al. 2007). It is of interest to mention that SigX also regulates the opdC (occD2) gene, as evidenced by ChIP-seq, RNA-seq and the presence of the SigX motif (Schulz et al. 2015), while opdP (occD3) expression has been proposed to depend on RpoN, although not demonstrated by ChIP-seq (Table 1, Fig. 7).
Phylogenetic relationships among OccD and OccK sequences from P. aeruginosa PAO1. The unrooted dendrogram was generated using neighbor-joining algorithm from evolutionary distances using Poisson correction. Bootstrap values correspond to 1000 pseudo-replicates.
OprQ (OccD6)
Despite a high level of similarity with OprD (OccD1) (>50%), OprQ (OccD6), contrary to OprD (OccD1), has no role in antibiotic resistance (Okamoto et al. 1999). Later on it was found that a P. aeruginosa mutant with an oprQ (occD6) gene disruption had a surprisingly stronger growth phenotype compared to the wild type in LB and in minimal M63 media while the mutant complemented with oprQ (occD6) in trans had growth characteristics undistinguishable from the wild type (Arhin and Boucher 2010). Interestingly, overproduction of oprQ (occD6) resulted in moderately increased sensitivity to some antibiotics (Arhin and Boucher 2010). The same authors showed that low magnesium increased the expression of oprQ (occD6) by a factor of 5 while a 13-fold increase was observed under low-iron conditions in minimal medium (Arhin and Boucher 2010). When planktonic P. aeruginosa PA14 cells are exposed for 30 min to anoxic conditions in CF sputum medium, a four-time upregulation of oprQ (occD6) was observed (Tata et al. 2016). In a gentamicin-resistant SCV of PAO1, oprQ (occD6) transcription is increased by a factor 4 compared to the wild type in late stationary phase (Wei et al. 2011). Jaouen et al. (2006) observed that a P. fluorescens oprQ (occD6) mutant could not anymore bind to the extracellular matrix fibronectin. The same observation could be made for the OprQ (OccD6) ortholog in P. aeruginosa since an oprQ (occD6) mutant could not bind to fibronectin anymore while a mutant complemented in trans with oprQ (occD6) had a restored binding capacity, especially after IPTG induction of the oprQ (occD6) expression (Arhin and Boucher 2010). This result suggests that OprQ (OccD6) is contributing to the human host colonization. Another interesting observation is that oprQ (occD6) disruption results in increased production of the phenazine pigment pyocyanin, which is known to contribute to virulence (Arhin and Boucher 2010). As already mentioned, an oprD (occD1) deletion mutant showed an increased expression of other porin genes, including oprQ (occD6) (Skurnik et al. 2013). Interestingly, a coregulation of the two porin genes oprD (occD1) and oprQ (occD6) has also recently been observed since both genes are overexpressed in a Hfq mutant of P. aeruginosa (Pusic et al. 2016). The two ECF sigma factors RpoS and SigX have been shown to bind the regulatory region upstream of oprQ (Fig. 7) (Schulz et al.2015).
OprE (OccK8)
The oprE (occK8) gene is transcribed as a single gene unit and codes for a 49.7 kDa porin first mentioned as being induced by anaerobic conditions (Yamano, Nishikawa and Komatsu 1993). It was later shown that deficiency in the alternative sigma factor RpoN resulted in non-expression of the porin both at transcription and protein levels under aerobic conditions, although it had no effect on the expression of oprE (occK8) when the cells were grown under anaerobic conditions (Yamano, Nishikawa and Komatsu 1998). Recently, the involvement of RpoN sigma factor in the regulation of oprE (occK8) has been confirmed via ChIP-seq analysis (Schulz et al. 2015) (Fig. 7) (Table 1). In a proteomic study, OprE (OccK8) production was found to be increased after exposition to alkanes, which makes sense because oxygen availability is reduced under these growth conditions (Hemamalini and Khare 2014). Interestingly, under the same conditions of alkanes exposure, the production of OprF and OprD (OccD1) is also reduced (Hemamalini and Khare 2014). A putative binding site for the LysR regulator OxyR has also been detected in the intergenic region upstream of oprE (occK8) by chromatin immunoprecipitation (Wei et al. 2012). Like OprD (OccD1), OprE (OccK8) is over-represented in OMVs from biofilm samples compared to the outer membrane of the corresponding bacteria at all three investigated time points (Park et al. 2015).
OpdK (OccK1)
OpdK (OccK1), which belongs to the OccK subfamily, is involved in the uptake of vanillate, and its structure has been determined (Fig. S1); it has 18 β-strands with a kidney-shaped central pore, and was found to form trimers in the outer membrane (Biswas et al. 2008; Cheneke, van den Berg and Movileanu 2011; Wang et al. 2012; Pothula and Kleinekathofer 2015). To the best of our knowledge, nothing is known about the regulation of opdK (occK1) expression.
OpdQ (OccK6)
The opdQ (occK6) gene has been shown before to be the only porin gene induced by the exposure of P. aeruginosa to the oral microflora (Duan et al. 2003). Later on its expression was found to be decreased by exposure to the QS molecules N-acyl-homoserine lactones and increased in a lasR mutant unable to sense these signal molecules (Schuster et al. 2003; Wagner et al. 2003).
Although its structure has been determined (Fig. S1), not much was known about OpdQ (OccK6) substrate specificity, until a recent interesting publication about its regulation involving the NarXL two-component system (Fowler and Hanson 2015) (Table 1). The same authors found that opdQ (occK6) is transcriptionally repressed under aerobic conditions while being induced by nitrate, which is dependent on the NarXL two-component system (Fowler and Hanson 2015). Five-prime rapid amplification of cDNA-ends (RACE) experiments allowed to determine the transcriptional start point of the opdQ (occK6) gene and putative –10 and –35 promoter sequences were determined as well as two regions forming stem-loop structures with one of them encompassing the –35 element (Fowler and Hanson 2015). Although transcription levels of opdQ (occK6) were lower in low oxygen conditions (2%), addition of nitrate in the same condition increased the level of transcription and resulted in increased OpdQ (OccK6) protein levels as well (Fowler and Hanson 2015). The NarXL two-component system has been previously determined to be involved in the response of P. aeruginosa to nitrate under low oxygen condition (Schreiber et al. 2007; Schobert and Jahn 2010; Trunk et al. 2010; He et al. 2014). Using a narL deletion mutant, Fowler and Hanson (2015) observed no change in expression of opdQ (occK6) by nitrate addition under ambient oxygen condition, but a strong decrease under micro aerobic condition (2% O2), confirming the involvement of NarL in the regulation of OpdQ (OccK6) production under anaerobic conditions in the presence of nitrate. The same authors suggested that OpdQ (OccK6) might serve as a port of entry for nitrate under low oxygen condition although it has been suggested in previous reports that OpdQ (OccK6) is a porin for carboxylates, including benzoate (Eren et al.2012; Liu et al. 2012a,b).
OpdT (OccD4)
OpdT (OccD4) is a tyrosine-specific porin (Tamber, Ochs and Hancock 2006). ZnO microparticles have been found to inhibit biofilm formation and the production of the virulence factor pyocyanin and of the signal molecule PQS (Lee et al. 2014) in a concentration that does not affect the planktonic growth. The same authors found that ZnO particles caused a higher expression of 34 genes, including the czc operon for Zn efflux system, an effect dependent on the CzcR and RhlR regulators. The inhibition of biofilm formation by ZnO microparticles was much less pronounced in the czcR and rhlR single mutants (Lee et al. 2014). Interestingly, the ZnO microparticles caused a decrease in the expression of the oprD (occD1) porin gene by a factor 8 while increasing the transcription of opdT (occD4) by a factor 14, results validated by quantitative RT-PCR (Lee et al. 2014). It is interesting to mention that the regulation of oprD (occD1) and opdT (occD4) by CzcR occurs in opposite directions, i.e. repression for oprD (occD1) and activation for opdT (occD4) (Dieppois et al. 2012; Lee et al. 2014) (Fig. 6, Table 1, Fig. S1).
OpdO (OccK3)
OpdO (OccK3) has been described to be involved in the uptake of pyroglutamate (Tamber, Ochs and Hancock 2006). Interestingly, OpdO (OccK3) is the most upregulated porin protein when the AmiE aliphatic amidase is overproduced in P. aeruginosa, which results in decreased virulence and reduced production of pyocyanin (Clamens et al. 2017).
INVOLVEMENT OF ECF SIGMA FACTORS IN PORIN GENE EXPRESSION
As already mentioned, several ECF sigma factors are involved in the expression of some porin genes. A comprehensive survey of 10 different Pseudomonas aeruginosa PA14 ECF sigma factors global transcriptional network (‘sigmulon’) has recently been published (Schulz et al. 2015). This study combined different approaches: ChIP-seq, RNA-seq and presence of a consensus DNA-binding motif (Table 1). After carefully analyzing their data, including in the supplementary file showing the results of a ChIP-seq analysis (the most stringent assay), we found that several porin genes are controlled by one or more ECF σ (Table 1 and Fig. 7). These data revealed that some ECF σ influence the expression of more than one porin gene, oprG, oprF, oprD (occD1), opdC (occD2) and oprE (occK8) for SigX, oprH, oprF, oprP (and/or oprO) and oprQ (occD6) for RpoS, oprD (occD1) and opdC (occD2) for FliA, and oprD (occD1) and oprE (occK8) for RpoN. Likewise, some porin genes are controlled by more than one ECF σ. This is the case for oprH (RpoS and FecI), oprF (AlgU, SigX and RpoS), oprD (occD1) (SigX, RpoN and FliA), oprQ (occD6) (SigX and RpoS) and oprE (occK8) (RpoN, FecI2 and SigX).
PORIN GENES CONSERVATION
A BlastP search (December 2016) for all porin proteins from PAO1 in all Pseudomonas aeruginosa complete genomes available in the pseudomonas database (www.pseudomonas.com) (Winsor et al. 2016) revealed a remarkable degree of conservation of these predicted proteins, suggesting that, at least within the P. aeruginosa species, the porin genes are part of the core genome. This is in contrast with a previous search for TonB-dependent receptors protein sequences, which revealed that some of them were not shared among the different strains available at the time (Cornelis and Bodilis 2009). There are however some exceptions, which are presented in Table 3. Among the porins present in all 60 P. aeruginosa sequenced complete genomes, 10 are fully conserved (OprF, OprG, OprH, OprE (OccK8), OprQ (OccD6), OpdC (OccD2), OpdB (OccD7), OpdF (OccK2) and OpdR (OccK11)). The percentage of amino acid sequence identity for porin orthologs is generally very high, always higher than 90%, with few exceptions (OpdI (OccD5), OpdR (OccK11) > 80%) in the case of the PA7 isolate, a highly antibiotic-resistant strain isolated in Argentina, which is well known to diverge from all the other P. aeruginosa representatives (Roy et al. 2010). The number of amino acids substitutions in PA7 porin proteins is also the highest, confirming its outlier character (Table S1). The number of amino acids substitutions in PA7 porin proteins is also the highest, confirming its outlier character (Table S1, Supporting Information). Not surprisingly, because of its involvement in carbapenem sensitivity, OprD (OccD1) is absent in 10 genomes (Table 3). The other frequently lost porins are OpdG (OccK9), which is absent from three genomes, OpdD (OccK7), also from three genomes, OpdP (OccD3) and OpdT (OccD4) which are both lost twice. If one excepts the OprD (OccD1) situation, there does not seem to be a correlation with the type of isolate and a specific porin gene loss. More intriguing is the loss of both OprO and OprP phosphate uptake porins in a keratitis strain (Stewart et al. 2011) and of the OprB and OprB2 glucose porins in an ovine mastitis strain although one can argue in this case that it reflects an adaptation to this specific niche (Wright et al. 2015). Interestingly, after 1 year in water, the PAO1 strain has been re-sequenced, and both OprD (OccD1) and OpdP (OccD3) were lost, suggesting that this strain should be deficient in the uptake of arginine as previously evidenced in the case of a double oprD (occD1) opdP (occD3) mutant (Tamber and Hancock 2006). Some porin genes are present in two copies: this is the case for opdD (occk7) in the VRFPA04 keratitis strain (Table 3) and, intriguingly, for oprF, in the two PA1 and PA1R strains, where the entire ppsA-estX-menG-cmaX-cfrX-cmpX-sigX-oprF locus is duplicated.
Porin protein sequences undetected in different P. aeruginosa genomes after a BlastP search on 60 complete genomes.
| Genome . | Origin . | Absent porins . | Phenotype/project . | Reference . | ||
|---|---|---|---|---|---|---|
| 19BR | Human Brazil | OprD (OccD1) | OpdG (OccK9) | Polymyxin R | Boyle et al. (2012) | |
| 213BR | Human Brazil | OprD (OccD1) | OpdG (OccK9) | Polymyxin R | Boyle et al. (2012) | |
| S86968 | Cancer | OpdG (OccK9) | PRJNA253624 | |||
| 39016 | Keratitis | OprP | OprO | OpdD (OccK7) | Stewart et al. (2011) | |
| B136-33 | Human diarrhea | OpdK (OccK1) | ||||
| DK2 | CF Denmark | OpdL (OccK4) | OpdN (OccK10) | Yang, Jelsbak and Molin (2011) | ||
| NCGM2S1 | MDR Japan | OprD (OccD1) | OpdD (OccK7) | Miyoshi-Akiyama et al. (2011) | ||
| PA7 | MDR Argentina | OpdD (OccK7) | Roy et al. (2010) | |||
| PA96 | M18 clone MDR | OpdO (OccK3) | Xiong et al. (2013) | |||
| PAO1H2O | PAO1-water | OprD (OccD1) | OpdP (OccD3) | PRJNA83447 | ||
| T63266 | Cancer | OpdP (OccD3) | PRJNA253624 | |||
| RP73 | CF Canada | OprD (OccD1) | OpdT (OccD4) | Jeukens et al. (2013) | ||
| F30658 | Cancer | OprD (OccD1) | OpdT (OccD4) | PRJNA253624 | ||
| F22031 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| F23197 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| F9670 | Cancer | OpdQ (OccK6) | ||||
| FRD1 | CF mucoid | OpdI (OccD5) | OpdJ (OccD8) | Lindsey et al. (2008) | ||
| PSE305 | Ovine mastitis | OprB | Wright et al. (2015) | |||
| W36662 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| VRFPA04 | Keratitis India | OprD (OccD1) | OpdH (OccK5) | Murugan et al. (2016) | ||
| Genome . | Origin . | Absent porins . | Phenotype/project . | Reference . | ||
|---|---|---|---|---|---|---|
| 19BR | Human Brazil | OprD (OccD1) | OpdG (OccK9) | Polymyxin R | Boyle et al. (2012) | |
| 213BR | Human Brazil | OprD (OccD1) | OpdG (OccK9) | Polymyxin R | Boyle et al. (2012) | |
| S86968 | Cancer | OpdG (OccK9) | PRJNA253624 | |||
| 39016 | Keratitis | OprP | OprO | OpdD (OccK7) | Stewart et al. (2011) | |
| B136-33 | Human diarrhea | OpdK (OccK1) | ||||
| DK2 | CF Denmark | OpdL (OccK4) | OpdN (OccK10) | Yang, Jelsbak and Molin (2011) | ||
| NCGM2S1 | MDR Japan | OprD (OccD1) | OpdD (OccK7) | Miyoshi-Akiyama et al. (2011) | ||
| PA7 | MDR Argentina | OpdD (OccK7) | Roy et al. (2010) | |||
| PA96 | M18 clone MDR | OpdO (OccK3) | Xiong et al. (2013) | |||
| PAO1H2O | PAO1-water | OprD (OccD1) | OpdP (OccD3) | PRJNA83447 | ||
| T63266 | Cancer | OpdP (OccD3) | PRJNA253624 | |||
| RP73 | CF Canada | OprD (OccD1) | OpdT (OccD4) | Jeukens et al. (2013) | ||
| F30658 | Cancer | OprD (OccD1) | OpdT (OccD4) | PRJNA253624 | ||
| F22031 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| F23197 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| F9670 | Cancer | OpdQ (OccK6) | ||||
| FRD1 | CF mucoid | OpdI (OccD5) | OpdJ (OccD8) | Lindsey et al. (2008) | ||
| PSE305 | Ovine mastitis | OprB | Wright et al. (2015) | |||
| W36662 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| VRFPA04 | Keratitis India | OprD (OccD1) | OpdH (OccK5) | Murugan et al. (2016) | ||
The porins absent in more than one genome are shaded in different tones of gray: OprD 10 genomes (dark gray and bold), OpdG and OpdD three genomes (gray and underlined), OpdP and OpdT two genomes (light gray).
Porin protein sequences undetected in different P. aeruginosa genomes after a BlastP search on 60 complete genomes.
| Genome . | Origin . | Absent porins . | Phenotype/project . | Reference . | ||
|---|---|---|---|---|---|---|
| 19BR | Human Brazil | OprD (OccD1) | OpdG (OccK9) | Polymyxin R | Boyle et al. (2012) | |
| 213BR | Human Brazil | OprD (OccD1) | OpdG (OccK9) | Polymyxin R | Boyle et al. (2012) | |
| S86968 | Cancer | OpdG (OccK9) | PRJNA253624 | |||
| 39016 | Keratitis | OprP | OprO | OpdD (OccK7) | Stewart et al. (2011) | |
| B136-33 | Human diarrhea | OpdK (OccK1) | ||||
| DK2 | CF Denmark | OpdL (OccK4) | OpdN (OccK10) | Yang, Jelsbak and Molin (2011) | ||
| NCGM2S1 | MDR Japan | OprD (OccD1) | OpdD (OccK7) | Miyoshi-Akiyama et al. (2011) | ||
| PA7 | MDR Argentina | OpdD (OccK7) | Roy et al. (2010) | |||
| PA96 | M18 clone MDR | OpdO (OccK3) | Xiong et al. (2013) | |||
| PAO1H2O | PAO1-water | OprD (OccD1) | OpdP (OccD3) | PRJNA83447 | ||
| T63266 | Cancer | OpdP (OccD3) | PRJNA253624 | |||
| RP73 | CF Canada | OprD (OccD1) | OpdT (OccD4) | Jeukens et al. (2013) | ||
| F30658 | Cancer | OprD (OccD1) | OpdT (OccD4) | PRJNA253624 | ||
| F22031 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| F23197 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| F9670 | Cancer | OpdQ (OccK6) | ||||
| FRD1 | CF mucoid | OpdI (OccD5) | OpdJ (OccD8) | Lindsey et al. (2008) | ||
| PSE305 | Ovine mastitis | OprB | Wright et al. (2015) | |||
| W36662 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| VRFPA04 | Keratitis India | OprD (OccD1) | OpdH (OccK5) | Murugan et al. (2016) | ||
| Genome . | Origin . | Absent porins . | Phenotype/project . | Reference . | ||
|---|---|---|---|---|---|---|
| 19BR | Human Brazil | OprD (OccD1) | OpdG (OccK9) | Polymyxin R | Boyle et al. (2012) | |
| 213BR | Human Brazil | OprD (OccD1) | OpdG (OccK9) | Polymyxin R | Boyle et al. (2012) | |
| S86968 | Cancer | OpdG (OccK9) | PRJNA253624 | |||
| 39016 | Keratitis | OprP | OprO | OpdD (OccK7) | Stewart et al. (2011) | |
| B136-33 | Human diarrhea | OpdK (OccK1) | ||||
| DK2 | CF Denmark | OpdL (OccK4) | OpdN (OccK10) | Yang, Jelsbak and Molin (2011) | ||
| NCGM2S1 | MDR Japan | OprD (OccD1) | OpdD (OccK7) | Miyoshi-Akiyama et al. (2011) | ||
| PA7 | MDR Argentina | OpdD (OccK7) | Roy et al. (2010) | |||
| PA96 | M18 clone MDR | OpdO (OccK3) | Xiong et al. (2013) | |||
| PAO1H2O | PAO1-water | OprD (OccD1) | OpdP (OccD3) | PRJNA83447 | ||
| T63266 | Cancer | OpdP (OccD3) | PRJNA253624 | |||
| RP73 | CF Canada | OprD (OccD1) | OpdT (OccD4) | Jeukens et al. (2013) | ||
| F30658 | Cancer | OprD (OccD1) | OpdT (OccD4) | PRJNA253624 | ||
| F22031 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| F23197 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| F9670 | Cancer | OpdQ (OccK6) | ||||
| FRD1 | CF mucoid | OpdI (OccD5) | OpdJ (OccD8) | Lindsey et al. (2008) | ||
| PSE305 | Ovine mastitis | OprB | Wright et al. (2015) | |||
| W36662 | Cancer | OprD (OccD1) | PRJNA253624 | |||
| VRFPA04 | Keratitis India | OprD (OccD1) | OpdH (OccK5) | Murugan et al. (2016) | ||
The porins absent in more than one genome are shaded in different tones of gray: OprD 10 genomes (dark gray and bold), OpdG and OpdD three genomes (gray and underlined), OpdP and OpdT two genomes (light gray).
CONCLUSIONS
From this last literature survey, it is apparent that a large amount of information has become available on the Pseudomonas aeruginosa porins, including some structure/function analysis, and data on their regulation. It shows that the major OprF porin is strongly involved in envelope integrity via interactions with peptidoglycan and other outer membrane proteins, including OprL and OprI. It also demonstrates the implication of OprF, and more recently OprH, in other functions, including adhesion and recognition of external cues from the host. One other important message is the complex regulation of the expression of porin genes (when known) although much more data need certainly to be obtained to get a better insight into their dynamic regulation which reflect their involvement in the high adaptability of P. aeruginosa to changing environmental conditions. Although this review did not attempt to explore the conservation of porin genes within the entire Pseudomonas genus, their high degree of conservation within the P. aeruginosa species is striking. It is our hope that this review will stimulate more research on these fascinating outer membrane proteins, their function and regulation, not only in P. aeruginosa, but also in other pseudomonads.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSRE online.
FUNDING
This work was supported by grants from the Communauté d'Agglomération d'Evreux, the Conseil Général de l'Eure, the European regional development fund (ERDF) and the Normandie Regional Council.
Conflict of interest. None declared.