Abstract
HRAS is regulated by two neighbouring quadruplex-forming GC-elements (hras-1 and hras-2), located upstream of the major transcription start sites (doi: 10.1093/nar/gku 5784). In this study we demonstrate that the C-rich strands of hras-1 and hras-2 fold into i-motif conformations (iMs) characterized under crowding conditions (PEG-300, 40% w/v) by semi-transitions at pH 6.3 and 6.7, respectively. Nondenaturing PAGE shows that the HRAS C-rich sequences migrate at both pH 5 and 7 as folded intramolecular structures. Chromatin immunoprecipitation shows that hnRNP A1 is associated under in vivo conditions to the GC-elements, while EMSA proves that hnRNP A1 binds tightly to the iMs. FRET and CD show that hnRNP A1 unfolds the iM structures upon binding. Furthermore, when hnRNP A1 is knocked out in T24 bladder cancer cells by a specific shRNA, the HRAS transcript level drops to 44 ± 5% of the control, suggesting that hnRNP A1 is necessary for gene activation. The sequestration by decoy oligonucleotides of the proteins (hnRNP A1 and others) binding to the HRAS iMs causes a significant inhibition of HRAS transcription. All these outcomes suggest that HRAS is regulated by a G-quadruplex/i-motif switch interacting with proteins that recognize non B-DNA conformations.
Similar content being viewed by others
Introduction
The HRAS oncogene encodes for a 21-kD GTP-ase conveying signals to the nucleus that stimulate cell proliferation1. In many tumours HRAS is mutated, normally in exon 1, codon 12, 13 or 61 and encodes for an altered protein which constitutively activates downstream pathways causing normal cells to become cancerous cells2. In previous works, we have demonstrated that HRAS is regulated by two neighbouring GC-rich elements that we called hras-1 (nt 432-464, A.N. J00277) and hras-2 (nt 509-530, A.N. J00277), located immediately upstream of the major transcription start sites (TSS’s), each capable of folding into a G-quadruplex structure3,4. By site-directed mutagenesis of the GC-elements, we found that the G-quadruplexes behave as transcription repressors3. Under normal conditions, hras-1 and hras-2 are folded into G-quadruplexes, thus locking the promoter into an inactive state characterized by a low transcription level3. Transcription is activated when the G-quadruplexes are unfolded and the G-elements transformed into canonical B-DNA forms. We found that MAZ, a zinc-finger transcription factor recognizing blocks of guanines, interacts with the promoter GC-elements under cellular conditions3. MAZ is an essential protein for gene expression, as it unfolds the HRAS G-quadruplexes and activates transcription3,4. Our data support a transcription model according to which the two neighbouring G-quadruplexes behave as a molecular switch that controls gene expression.
In the present work we interrogated if the complementary C-rich strands of hras-1 and hras-2 (namely hras-1Y and hras-2Y) fold into the well known iM conformation5,6,7,8,9,10,11,12,13,14,15. We found that hras-1Y and hras-2Y assume the iM conformation under slightly acidic conditions, which are close to neutrality in the presence of a crowding agent, for example PEG-30016. We also discovered that the HRAS iMs are recognized by nuclear proteins, including nuclear factor hnRNP A1. This protein, which shows a binding preference for cytosines, unfolds the iM conformation of the HRAS sequences. When hnRNP A1 was knocked out in T24 bladder cancer cells by a specific shRNA, the level of HRAS transcript also dropped to 44 ± 5% of the control. Together, our data provide evidence that hnRNP A1, with its unfolding activity against the iM, is an essential factor for the activation of HRAS. Indeed, when hnRNP A1 was sequestered by decoy oligonucleotides mimicking the iM, HRAS transcription was significantly downregulated. The outcome of this work support the notion that HRAS expression is regulated by a G-quadruplex/iM switch that is controlled by proteins.
Results and Discussion
The sequence of the HRAS promoter immediately upstream of the major transcription start sites is reported in Fig. 1A. It contains two GC-rich elements, hras-1 and hras-2, composed of blocks of guanines and capable of folding into G-quadruplex structures. In previous works we have demonstrated that these sequences behave as a regulatory switch controlling gene expression3,4. Such a mechanism has been proposed for other relevant oncogenes including KRAS17,18, CKIT19,20 and CMYC21,22. A couple of comprehensive reviews on this subject have been reported23,24. In this work we have focused on the complementary C-rich strands hras-1Y and hras-2Y and have investigated if they fold into stable iMs.
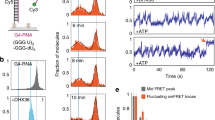
(A) Sequences of the GC-rich elements located in the HRAS promoter upstream of major TSS’s; (B,C) Circular dichroism titrations of hras-1Y and hras-2Y (3 μM, 1 cm pathlength cell) in 50 mM Tris-acetate, 50 mM KCl, 40% PEG-300 and pH from 4.5 to 8; (D) Ellipticity (287 nm) versus pH curves for hras-1Y and hras-2Y in the presence and absence of PEG-300; (E) Determination of number of protons picked up by hras-1Y and hras-2Y upon folding into the iM.
iM formation by the HRAS C-rich sequences
To find out if hras-1Y and hras-2Y can assume the iM conformation, we performed circular dichroism (CD) experiments as a function of pH, in 50 mM KCl, 50 mM Tris-acetate, 25 °C. To mimic the crowding conditions of the cell, we analysed the sequences both in the presence and absence of 40% (w/v) PEG-30016. Typical CD titrations are shown in Fig. 1B,C. It can be seen that the spectra of hras-1Y and hras-2Y change dramatically as the pH is gradually decreased from 8 to 4.5. Under acidic conditions (pH 5) both sequences exhibit the characteristic enhanced ellipticity at ~287 nm of a classical iM10,12,25,26,27, while at pH 8 the sequences exhibit a much lower ellipticity, shifted at ~285 nm. By plotting the 287-nm ellipticity as a function of pH, we obtained for each sequence, in the presence or absence of PEG-300, sigmoidal curves reflecting iM formation (Fig. 1D). The crowding agent drives the folding at higher pH values: the semi-transition of hras-1Y increases from pH 5.9 to 6.3, whereas that of hras-2Y increases from pH 6.2 to 6.7. These plots suggest that the iMs are stable in a slightly acidic medium. However, under cellular conditions the iM can be stabilized by: (i) transcriptionally induced DNA superhelicity28,29; (ii) more effective cellular crowding conditions30,31; (iii) an increased intracellular acidity generated by an increase of the glucose-lactate flux32,33.
As the ellipticity-versus-pH curves are reversible, we evaluated the number of protons involved in the folding of hras-1Y and hras-2Y, by considering the following equilibria:


where equilibrium 1 takes into consideration the fact that the unfolded C-rich sequence, U, may bind m protons before folding into the i-motif (as the pKa of cytosine is ∼4.434, at pH 5 about 3 cytosines out of 12 are expected to be protonated); U*·mH+ is the unfolded sequence with m bound protons. The bases in U*·mH+ are assumed to be unstacked, so the formation of this species is accompanied by a negligible change in the CD spectrum35. In contrast, upon folding into i-motif F, U*·mH+ assumes in a cooperative manner other n protons to form (n+m) CH+:C base pairs that are stacked in the structure. A significant CD spectral change is expected for the formation of the iM8,10 (equilibrium 2), whose equilibrium constant is KD = ([F·(m+n)H+]/([U*·mH+][H+]n) (3). From the CD plots of Fig. 1B,C, we determined for each sequence the fraction of folded and unfolded species and ratio [F(n+m)H+]/[ U*·mH+]. By plotting log [F(n+m)H+]/[ U*·mH+] versus pH, we obtained a straight line whose slope is n (Fig. 1E). We obtained values of n ∼ 4 and n ∼ 2 for hras-1Y and hras-2Y, respectively. This suggests that when hras-1Y and hras-2Y fold into the iM, they assume 4 and 2 protons, respectively, which agrees with the fact that the sequences are partially protonated before folding. A similar behaviour has been previously observed for the formation of the i-motif by (C3TA2)414.
PAGE and analyses of melting curves
The intramolecular iM, being a folded structure, migrates in a polyacrylamide gel faster than an unfolded oligonucleotide of the same length. We analysed hras-1Y and hras-2Y by PAGE under different pH conditions. The mobility of the two C-rich sequences was compared with that of hras-1Y variants: ODN-1 (unable to form any structure); ODN-2 (forming an iM with 4 CH+:C) and ODN-3 (forming a stable W.C. hairpin) (Fig. 2A,B) (Methods). Under denaturing conditions (7 M urea), the 27-mer oligonucleotides (hras-1Y and variants) exhibited the same mobility, with the exception of ODN-3 that forms a hairpin even in the presence of 7 M urea. Sequence hras-2Y, being embedded in a 36-mer oligonucleotide, migrates slowly (Methods). In contrast, under native conditions at pH 5, hras-1Y, that assumes the iM according to CD, migrates with a sharp band faster than that of ODN-1. Interestingly, when the hras-1Y sequence is modified to fold into a hairpin stabilized by a stem of 8 W.C. bps (ODN-3) (S1), it migrates even quicker than the iM. This is because the 6 positive charges of iM reduce its negative charge density. The folding of hras-1Y into the iM is quite fast, as the mobility does not change when the sample is heated before loading. Variant ODN-2, forming an iM with 4 positive charges (S1), migrates slightly faster than hras-1Y, as expected. At pH 5, the 36-mer oligonucleotide containing sequence hras-2Y migrates quicker than unstructured 27-mer ODN-1, as it folds into the iM (see CD). At pH 7, both hras-1Y and hras-2Y still migrate faster than ODN-1, indicating that even at neutral pH the sequences are folded. However, they migrate either with a smeared band (hras-1Y) or with two bands (hras-2Y), suggesting that more than one folded structure is formed: most likely iM-like structures stabilized by C:C and CH+:C bps. It is also possible that at pH 7, the iM-like structure is in equilibrium with a flexible hairpin (Fig. 2C, S1).
(A) PAGE of hras-1Y and hras-2Y and variants ODN-1, ODN-2 and ODN-3 in denaturing conditions (7 M urea); native conditions at pH 5 and 7. The oligonucleotide sequences are reported in Methods. The electrophoresis was run at 20 °C, the bands stained with stains-all. Δ = heated 10 min 90 °C; (B) CH+:C base pair; (C) Putative equilibrium between iMs involving different protonation levels and a flexible hairpin formed by hras-1Y at pH ≥ 7.
We have also examined the thermal stability of the iMs by CD- and FRET-melting experiments. Typical CD-melting profiles for hras-1Y and hras-2Y are reported in Fig. 3A,B. By heating (20 → 85 °C) and then cooling (85 → 20 °C) the DNA solutions in 50 mM sodium cacodylate, pH 5, 50 mM KCl, the folded/unfolded transitions showed to be cooperative and reversible, as previously found for C-rich oligonucleotides under similar experimental conditions10,12,13,14. The melting in the pH range between 5 and 7 was examined by FRET experiments, using oligonucleotides end-labelled with ATTO-488 (5′-end) and TAMRA (3′-end) (Fig. 3C,D). The TM of both hras-1Y and hras-2Y decreased with increasing pH: hras-1Y, 55.9, 52.1, 47.6, 44.8, 41.3 °C at pH 5, 5.5, 6, 6.5, 7, respectively; hras-2Y, 65.5, 58.7, 55.1, 53.8, 53.1 °C at pH 5, 5.5, 6, 6.5, 7. At pH values > 5, the TM of the iMs decreases as the structure is probably stabilized by a number of CH+:C < 6. It is indeed reasonable to assume that when the medium is not sufficiently acidic, the iM is stabilized by both C:C and CH+:C base pairs36,37. iM structures of different protonation levels may coexist in solution. Mechanical stability experiments with the ILPR C-rich sequence showed that partially folded iM-like species are in equilibrium with fully folded iM at neutral pH36. The higher TM of hras-2Y is likely due to the additional CH+:C bp that stabilizes the iM. The two structures have similar rupture forces and sufficient stability to stall RNA polymerase36. Yang and Rodgers have reported that the energy of C:C is about 1/3 of that of CH+:C38. Sequence hras-2Y shows a behaviour similar to hras-1Y, with the difference that at pH 5 it shows a higher TM, 65.5 °C (Fig. 3D).
(A,B) CD melting curves of 3 μM hras-1Y and hras-2Y in 50 mM sodium cacodylate, pH 5, 50 mM KCl. Denaturing curve (20 → 85 °C), renaturing curve (85 → 20 °C); (C,D) Fraction of iM versus T curves obtained from FRET-melting experiment (0.3 °C/min), in 50 mM sodium cacodylate, pH 5 to 7, 50 mM KCl. The curves of hras-1Y and hras-2Y at pH 5 are fully reversible. FRET-melting gives TM values ∼3 °C higher than CD-melting values, due to the presence of the fluorophores in the oligonucleotides analysed by FRET.
We evaluated the thermodynamics of the folding transitions according to a two-state model. This was done at pH 5, where the two sequences fold into only one structure, as shown by PAGE. From the CD- and FRET-melting curves at pH 5, we obtained the following average thermodynamic data (±10%): ΔH = 252 kJ/mol and ΔS = −770 J/mol K and ΔG = −17 kJ/mol for hras-1Y; ΔH = 323 kJ/mol and ΔS = −950 J/mol K and ΔG = −27 kJ/mol for hras-2Y. Assuming that the breaking of a CH+:C bp needs approximately 46 ± 4 kJ/mol12, the number of CH+:C broken by the thermal disruption of hras-1Y is ~6, of hras-2Y is ~7, in accord with the number of expected protons that should bind to the sequences at pH near pKa10,12. At pH 5, nearly half of the cytosines is protonated and the sequences are completely folded into iM, showing the highest stability.
The HRAS iMs are recognized by hnRNP A1
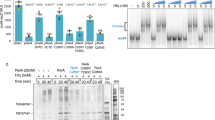
As the iM-forming sequences overlap critical GC-elements immediately upstream of TSS, we interrogated if these unusual structures are recognized by nuclear proteins. Previous studies have reported that DNA sequences composed by runs of cytosines such as the C-repeats in the telomeres and in the CMYC promoter are recognized by proteins of the heterogeneous nuclear riboproteins family (hnRNP)39,40 Moreover, Hurley and co-workers recently reported that the iM formed in the BCL2 promoter interacts with hnRNP LL6. Specific binding of heterogeneous ribonuclear proteins to C-rich DNA sequences is also supported by previous work, according to which hnRNP A1 binds to the GC-element of KRAS17, which shows a high sequence/functional homology with the HRAS GC-elements. HnRNP A1 is one of the most abundant nuclear proteins of eukaryotic cells that regulates several aspects of mRNA biogenesis41. As it is over-expressed in a variety of cancers41,42, we wondered if this protein plays a role in the promoter of the HRAS oncogene, in the region where the iM can potentially be formed. To address this question, we first investigated by chromatin immunoprecipitation (ChIP) if in HRAS-mutant T24 bladder cancer cells, hnRNP A1 is associated to the GC-elements under in vivo conditions. The occupancy of hras-1 and hras-2 (located 6 bp upstream of first TSS) by hnRNP A1 was compared with the occupancy of a reference GC-rich sequence unable to fold into a non-B DNA structure (located 870 bp downstream from first TSS). A typical ChIP is shown in Fig. 4A. We found that the occupancy of hras-1 and hras-2 by hnRNP A1 was, respectively, ~6- and ~5-fold higher than the occupancy by IgG (negative control). As hras-1 and hras-2 are located in the region of the major transcription start sites, they show a significant occupancy by RNA Pol II: ~4-fold higher than the IgG signal. In contrast, the reference sequence showed almost no occupancy by any of the proteins considered. The ChIP data provided strong evidence that hnRNP A1, under in vivo conditions, is indeed associated to the critical GC-elements of the HRAS promoter. However, ChIP data do not provide information about the conformation of the GC-elements interacting with hnRNP A1. To know if the nuclear factor recognizes the iM, we performed EMSA at pH 5.5 and 20 °C of mixtures composed by hras-1Y or hras-2Y and recombinant hnRNP A1, which was produced with a high degree of purity (Fig. 4B,C). It can be seen that at pH 5.5 hras-1Y and hras-2Y (which are in the folded iM conformation) form with hnRNP A1 a retarded band due to a 1:1 DNA-protein complex. In the presence of 4 μg hnRNP A1, the iM is completely bound to the protein (Fig. 4B). With higher protein amounts, a second retarded band of much weaker intensity, probably a 1:2 complex, can be seen in the gel. When hnRNP A1 was thermally denatured before being added to the iM, the DNA-protein complex was abrogated and the iM migrated as a free molecule. As a further control, we used an unspecific protein like BSA and we found that it did not bind to the iM, as expected (Fig. 4C). When the iMs was destabilized by replacing 4 C with 4 T (hras-1Y(m)) (see Methods), the binding was significantly attenuated, suggesting that the iM conformation is essential for optimal hnRNP A1 binding.
(A) ChIP experiment to determine the occupancy of hras-1, hras-2 and control sequence (870 bp downstream from TSS) by hnRNP A1. Histograms shows the relative occupancy of hras-1 and hras-2 by hnRNP A1, RNA Pol II (positive control) and IgG (negative control). Data have been normalized by IgG signal; (B) EMSA of 32P-labelled hras-1Y and hras-2Y in 50 mM Tris-acetate pH 5.5, 50 mM KCl, incubated 40 min at room temperature with increasing amounts of recombinant hnRNP A1 (0–12 μg). Lane (Δ,A1) indicates the iM incubated 40 min at room temperature, with denatured hnRNPA1 in binding buffer (see Methods); (C) EMSA at pH 5.5 of hras-1Y with BSA or denatured hnRNP A1 and EMSA of hras-1Y (m) with hnRNP A1; ss = single-stranded oligonucleotide; 1:1 and 1:2 DNA-protein complexes.
hnRNP A1 unfolds the HRAS i-motif
In a previous work we have found that the HRAS promoter is highly active when the GC-elements are unfolded in the double-stranded conformation3,4. We now asked if the binding of hnRNP A1 to the iM involves the unfolding of this non B-DNA structure. To this purpose, we performed FRET experiments with hras-1Y tagged with ATTO-488 (donor) and TAMRA (acceptor) in 50 mM sodium cacodylate, pH 5.5, 50 mM KCl. By exciting the donor at 480 nm, both donor (520 nm) and acceptor (580 nm) emit fluorescence, as a result of FRET between the fluorophores (Fig. 5A). When hras-1Y is folded into the iM, the energy transfer, ET, between the two fluorophores is 0.77 and their end-to-end distance is ∼40 Å (S2). The effect of hnRNP A1 on the iM was investigated by incubating protein and DNA for 1.5 h and measuring the fluorescence between 500 and 650 nm, upon donor excitation at 480 nm. It can be seen that hnRNP A1 causes a dramatic increase of the donor emission, accompanied by a decrease of ET as a function of r (r = [protein]/[iM]), in a dose-dependent manner, from 0.77 (r = 0) to 0.18 (r = 7) (inset). This means that the end-to-end distance in the iM increases from ~40 to ~64 Å, suggesting that upon binding to the protein, hras-1Y goes through a conformational change. When hras-1Y is in the duplex conformation, the fluorophores are separated by ~86 Å (26 × 0.33 = 86 Å, where 0.33 Å is the vertical rise per bp), as a 27-mer duplex behaves as an extended rigid rod. It follows that the iM bound to hnRNP A1 is not fully extended as in the duplex. As a control we used an unspecific protein as BSA and heated hnRNP A1 before incubation with the iM (20 min, 95 °C). In both cases the fluorescence of the donor did not increase, as expected (Fig. 5A).
(A) FRET spectra of 200 nM hras-1Y treated with increasing amounts of purified hnRNP A1 at pH 5.5, 50 mM sodium cacodylate, 50 mM KCl. As a control BSA and denatured hnRNP A1 (A1 Δ) have been used. Note that hnRNP A1 causes a dramatic increase of the 520 nm donor emission. The emission spectra of hras-1Y hybridized to its complementary strand to yield the duplex is reported. Insight shows the energy transfer (ET) between donor-acceptor as a function of hnRNP A1 concentrations; (B) FRET-melting of hras-1Y incubated with increasing amounts of hnRNP A1 (r = 1–7). The protein abrogates the melting profiles; (C) Cartoon showing the melting of hras-1Y bound to its complementary strand or to hnRNP A1 (r = 1–7); (D) –dRFU/dT versus T curves of hras-1Y alone, hras-1Y+protein, hras-1Y in duplex, i.e. hybridized to its complementary strand.
To further support the finding that hnRNP A1 disrupts the iM, we carried out FRET-melting experiments, reasoning that the iM would not give its typical melting profile when bound to hnRNP A1. Fig. 5B shows that hras-1Y iM in 50 mM sodium cacodylate, pH 5.5, 50 mM KCl has a TM ~ 50 °C. In the presence of 4 equivalents (r = 4) of BSA or denatured hnRNP A1, the melting profile of hras-1Y did not change, as expected. In contrast, when the hras-1Y iM was incubated with increasing amounts of native hnRNP A1 (r = 1–7), its melting profile strongly changed, in keeping with the fact that hras-1Y bound to the protein is not in the iM conformation. It is worth noting that free hras-1Y melts with an increasing sigmoidal curve, whereas hras-1Y bound to hnRNP A1 melts with a decreasing sigmoidal curve (Fig. 5B). A melting profile similar to that of the hras-1Y:hnRNP A1 complex is obtained with hras-1Y in the duplex conformation with its complementary strand, where the two fluorophores are separated by ~86 Å. Upon melting, the duplex releases the hras-1Y strand which, thanks to its flexibility, will have an end-to-end distance <86 Å (Fig. 5C). This results in a decreasing sigmoidal melting curve and thus in a –dRFU/dT versus T curve marked by a positive peak. In the same way, hras-1Y bound to hnRNP A1 is more extended than when it is free. Therefore, also the complex gives a decreasing melting curve and a first derivative curve with a positive peak at ∼50 °C. The melting of free hras-1Y folded in the iM gives an increasing melting curve and a –dRFU/dT versus T curve with a negative peak (Fig. 5D). Summing up, both FRET titrations and melting provide strong evidence that hnRNP A1 unfolds the HRAS iM.
We have also analysed the effect of hnRNP A1 on the hras-2Y iM. We found that the protein unfolds the iM of hras-2Y at higher r values, as the iM formed by hras-2Y has a higher stability than the hras-1Y iM (58.7 versus 52.1 °C, at pH 5.5) (S3).
The effect of hnRNP A1 on the HRAS iMs was also investigated by CD (Fig. 6). At pH 5.5, hras-1Y and hras-2Y show the typical strong ellipticity at 287 nm of the iM conformation. When the iMs are thermally denatured, the positive 287 nm ellipticity drops dramatically. This is a hallmark of the transformation of iM into ssDNA. A similar spectral change was obtained when we added increasing amounts of hnRNP A1 (r = 1, 2, 3, 4) to the iMs. HnRNP A1 causes a progressive reduction of the 287 nm ellipticity, indicating that the iM structures are unfolded by the protein. As already observed with the FRET experiments, the unfolding effect is stronger with hras-1Y than with hras-2Y, owing to the different stability of the two iMs.
Insights into the binding of hnRNP A1 to the hras-1Y iM
Clues to the binding mode of hnRNP A1 to the iM can be obtained from the co-crystal structure of UP1 (the N-terminus of hnRNP A1 with DNA binding activity) and the human telomeric sequence d(TTAGGG)243. The co-crystal shows that a protein dimer binds to two single-stranded strands, in the antiparallel orientation. As the two binding domains (RRM1 and RRM2) within each protein molecule are also antiparallel, the 5′ → 3′ polarity of ssDNA with respect to the RRM orientation is the same for each RRM. The two lateral loops of the iM may (after a minor adjustment) provide suitable binding sites for hnRNP A1, as they are antiparallel and separated by ~15–20 Å. So, the iM’s main function should be to provide a rigid chemical frame displaying two lateral loops with the precise nucleic acid directionality with respect to the RRM orientation. In other words, the iM structure should offer a kinetic advantage to the binding of hnRNP A1 (indeed, when the iM is disrupted, the binding is strongly attenuated, Fig. 4C). If we assume that the protein binds to the lateral loops, it can form either a 1:1 or a 1:2 complex, depending whether one or two protein molecules bind to the iM. The equilibria occurring in solution are: P + iM = P•iM (4); P + P•iM = (P)2•iM (5), where P is hnRNP A1. In the presence of a large excess of P compared to iM (1:100), both equilibria shift to the right forming complex 1:2. In contrast, with less P (ratio 1:50) equilibrium (5) does not shift to the right and only complex 1:1 is formed (Fig. 7A, lanes 1–3). In addition, as hnRNP A1 unfolds the iM, its binding depends on temperature: at 0 °C only complex 1:1 is formed probably because it requires a partial unfolding of iM, at 37 °C complex 1:2 is favoured as it requires a complete opening of the iM (S4). To support the binding of the protein to the lateral loops we performed the following competition experiment. As the two lateral loops of the hras-1Y iM are separated by 10 nt, complex 1:2 should be competed by an oligonucleotide containing the two lateral-loop binding sites separated by a spacer of 10 nt (with a 10 nt spacer the competitor assumes a U-shape so that two antiparallel binding sites can interact with the protein RRMs). Moreover, if the spacer of the competitor is reduced to 8, 6, 4 and 2 nt, its capacity to compete with the formation of the 1:2 complex should gradually become weaker. To test this hypothesis we designed the competitors shown in Fig. 7C. When the competitors (150-fold in excess over iM) were incubated with the iM and hnRNP A1 (100-fold over iM), we found that the best competitor was the oligomer containing a spacer of 10 nt, which corresponds exactly to the distance of the lateral-loop binding sites in the wild-type hras-1Y iM. These data support the binding of hnRNP A1 to the lateral loops of the iM, as observed for hnRNP LL and BCL2 iM6. As stated above, the iM facilitates the initial binding to hnRNP A1. Then, after the iM is unfolded, the protein should bind more stably to the iM sequence.
(A) 5% PAGE, lanes 4–8 show how the binding of hnRNP A1 to hras-1Y–dy781 (Methods) (ratio 1:100) at pH 5.5 is competed by oligonucleotides 2-nt - 10-nt (150-fold over iM). Lanes 1–3 shows the binding of iM to hnRNP A1 at ratios 1:50, 1:75, 1:100. Experimental conditions: DNA and protein incubated for 40 min at 37 °C, pH 5.5 before 5% PAGE analysis; (B) structure of the iM with the two lateral loops to which the protein is expected to bind; (C) sequences of the oligonucleotide competitors 2-nt, 4-nt, 6-nt, 8-nt, 10-nt.
HnRNP A1 knockout results in downregulation of HRAS transcription in human bladder cancer cells.
As hnRNP A1 binds to the critical GC-elements of the HRAS promoter, we asked if the protein plays a role in transcription. We therefore evaluated the effect on transcription of knocking out hnRNP A1 by shRNA. First, we determined by quantitative real-time PCR the efficiency of shRNA to knock out hnRNP A1, finding that hnRNP A1 mRNA (normalized by the transcripts of β2-microglobulin and hypoxanthine guanine phosphoribosyltransferase, HPRT) was reduced to 29 ± 8% of the control (cells untreated or treated with a non-specific shRNAC), 48 h after treatment (Fig. 8A). In the same cells we also measured the level of HRAS mRNA finding that when hnRNP A1 is knocked out, the HRAS transcription is also down-regulated to 44 ± 5% of control. This suggests that hnRNP A1 is an essential factor for transcription, as previously reported41. Further evidence that the proteins recognizing the iMs of HRAS are essential for transcription was obtained with decoy oligonucleotides mimicking hras-1Y iM. These molecules, once introduced in the cells, should sequester the proteins (hnRNP A1 included) recognizing the C-rich strand of the HRAS GC-elements. To increase their nuclease resistance, the decoy oligonucleotides, namely 5291–5294 (see Methods), have been designed with unlocked nucleic acid (UNA) modifications (Fig. 8B) (S5)44,45. The capacity of the UNA-modified oligonucleotides to inhibit HRAS transcription was investigated by quantitative real-time PCR. T24 bladder cancer cells were transfected with the decoy oligonucleotides as well as with wild-type hras-1Y, using as transfecting agent jet-PEI46. After an incubation of 24 h, the total cellular RNA was extracted and the amount of HRAS mRNA relative to the housekeeping HPRT mRNA was evaluated by qRT-PCR (Fig. 8C). The results showed that oligonucleotides 5292 and 5293 reduced HRAS mRNA to ~50% of the control (untreated cells). We also examined by electrophoresis the nuclease resistance of the decoy oligonucleotides (Fig. 8D). The oligonucleotides were incubated in cell cultured medium containing 10% fetal bovine serum at 37 °C, pH 5.5 for 0, 18, 24 and 48 h. While hras-1Y was quickly degraded, the UNA-modified oligonucleotides, in particular 5292 and 5293, showed a remarkable stability, as the fraction of unbroken oligonucleotide was >0.5, after 48 h of incubation. Interestingly, the enhanced activity shown by these compounds correlates nicely with their higher stability in serum.
(A) Real-time determination of hnRNP A1 and HRAS mRNAs after knocking down hnRNP A1 in T24 bladder cancer cells with a specific shRNA. When hnRNP A1 is knocked down, HRAS mRNA is downregulated. P < 0.05 (*); (B) UNA modification introduced in the decoy oligonucleotides; (C) Level of HRAS mRNA in T24 cells treated with 200 nM hras-1Y or UNA-modified analogues. Total RNA was extracted 24 h after oligonucleotide transfection, retro-transcribed and subjected to real time amplification. HRAS mRNA expression is normalized with housekeeping gene HPRT. The percentage of residual HRAS mRNA compared to HPRT mRNA in each sample is reported. P < 0.05 (*); (D) Resistance in fetal serum of hras-1Y and UNA-modified analogues. Oligonucleotides have been incubated in serum for 0, 18, 24, 48 and 72 h at 37 °C. After incubation the samples have been run in denaturing PAGE, 7 M urea, 55 °C. The gels were stained with “stains all”.
Conclusion
We have demonstrated that two neighbouring GC-rich elements controlling HRAS expression can form non B-DNA iM structures, which are stable under near-physiological conditions. These unusual DNA structures are recognized by hnRNP A1, one of the most abundant nuclear proteins involved in the biogenesis of RNA. We have discovered that hnRNP A1 has a clear unfolding activity against the iM. As the knockout of hnRNP A1 by shRNA in T24 bladder cancer cells results in the inhibition of HRAS, hnRNP A1 behaves as an activating transcription factor. Our data, together with those of Hurley and co-workers, who showed that hnRNP LL binds to the iM of the BCL2 promoter and activates transcription6, provide the first evidence that non B-DNA iM structures are recognized by nuclear proteins.
The proteins of the hnRNP family have been associated with the promoter of several genes where they are supposed to participate in the transcription regulation mechanisms, although their exact role is not yet fully understood. Some of them recognize C-rich sequences in the promoters of CMYC (hnRNP K)39,40, BCL2 (hnRNP LL)6 and HRAS (hnRNP A1) (present study). These proteins seem to have a complex binding capacity, as hnRNP A1 is also able to bind to G-quadruplex DNA structures in KRAS17 and telomeres47. Remarkably, this type of binding is also associated with the disruption of G-quadruplex structures17.
Recent mechanical folding/unfolding experiments showed that G-quadruplex and iM are mutually exclusive within the same double-stranded tract48. However, whether this also holds under in vivo conditions, where double stranded DNA is exposed to negative superhelicity and located in a molecular crowding environment, has not yet been demonstrated. It is possible that both G-quadruplex and iM are extruded from each double-stranded GC-element, in the same way as two opposing hairpins (a cruciform) are extruded from a palindromic sequence. HRAS could therefore be regulated by a G-quadruplex/iM switch that represses transcription when the structural elements are in the folded conformation. Transcription will be activated when hnRNP A1 and MAZ, which recognize the HRAS G-quadruplexes3, bind to the iM and G-quadruplex, respectively and then to other proteins of the transcriptional activator complex. These non B-DNA structures provide a mechanism for the control of gene expression at a different level than duplex, involving proteins recognizing these unusual structures that play a central role in gene regulation.
Methods
Oligonucleotides and hnRNP A1
The oligonucleotides used in this study have been obtained from Microsynth AG (Switzerland) and Eurofins Genomics (Germany):
5′-CGCCCGTGCCCTGCGCCCGCAACCCGA (hras-1Y)
5′-ACCGCGCGCCCCCGCCCCCGCCCCGCCCCGGCCTCG (hras-2Y)
5′-ATTO-CGCCGCCCGTGCCCTGCGCCCGCAACCCGAGC-TAMRA (A-hras-1Y-T)
5′-ATTO-CGC GCC CCC GCC CCC GCC CCG CCC C -TAMRA (A-hras-2Y-T)
5′-TCG GGT TGC GGG CGC AGG GCA CGG GCG (hras-1R)
5′-CGG GGC GGG GCG GGG GCG GGG GCG (hras-2R)
5′- CGC TCG TGC TCT GCG CTC GCA ACT CGA (hras-1Ym)
5′- TTTTTGTGTTTTTTTTTGCAATTTTT (ODN-1)
5′-CGTCCGTGTCCTGCGTCCGCAATCCGA (ODN-2)
5′-CGCCCGTGCCCTGCGCCCGCAGGGCGA (ODN-3)
5′-Dy 781-TTTTTTTCGCCCGTGCCCGTCGCCCGCAACCCGATTTTTTT-3′ (hras-1Y-dy 781). The oligonucleotides with UNA modifications have been synthesized in solid phase as previously described44,45: 5′-CGCCCGTGCCCTGuCGCCCGCuAACCCGuA (5291); 5′- CGCCCGTGCCCuUGCGCCCuGCAACCCGuA (5292); CGCCCGuUGCCCTGuCGCC-CGCuAACCCGuA (5293) and 5′-CGCCCGuUGCCCuUGCGCCCuGCAACCCGuA (5294) where uC, uU, uG, uA are unlocked nucleic acid nucleotides.
Recombinant hnRNP A1 tagged to GST was obtained with a high degree of purity as previously described49 (S6).
Chromatin immunoprecipitation
T24 urinary bladder cancer cells (1.2 × 106) were cultured overnight in 6-cm diameter plates up to about 80% confluency and fixed in 1% formaldehyde in PBS for 5 minutes at room temperature to crosslink proteins to DNA. Chromatin immunoprecipitation assays were performed using the ChIP-ITTM Express kit (Active Motif, Rixensart, Belgium). Details are reported in S7.
ShRNA transfection, RNA extraction and real-time PCR
T24 cells were plated in 96-well plate (104 cells/well). After 1 day we transfected the cells with hnRNP A1-specific (sc-35576-SH) and control shRNA (sc-108066) (Santa Cruz, Dallas, USA) using as transfectant agent jetPEITM (Polyplus, NY, USA). After 48 h, RNA was extracted by using iScript TM RT-qPCR sample preparation reagent (BioRad, USA).
For cDNA synthesis, 1.25 μl of RNA was heated at 70 °C and placed in ice. The solution was added with 7.5 μl of a mix containing (final concentrations) 1 × buffer; 0.01 M DTT (Invitrogen); 1.6 μM primer dT [MWG Biotech, Ebersberg, Germany; d(T)16]; 1.6 μM random primers; 0.4 mM dNTPs solution containing equimolar amounts of dATP, dCTP, dGTP and dTTP (Euroclone, Pavia, Italy); 0.8 U/μl RNAse OUT; 8 U/μl of M-MLV reverse transcriptase (Life Technologies, Monza, Italy). The reactions were incubated for 1 h at 37 °C and stopped with heating at 95 °C for 5 min. As a negative control the reverse transcription reaction was performed with a sample containing DEPC water.
Real-time PCR multiplex reactions were performed with 1xKapa Probe fast qPCR kit for HRAS and housekeeping genes hypoxanthine-guanine phosphoribosyltransferase (HPRT) and β2-microglobulin, 2.2 μl of cDNA and primers/probes at the concentrations specified in S1. The PCR cycle was: 3 min at 95 °C, 50 cycles 10 s at 95 °C, 60 s at 58 °C. Real-time PCR amplification of hnRNP A1 was performed with 1 × Kapa Sybr Fast BioRad iCycle qPCR kit (KAPA Biosystems, Wilmington, MA, USA), 300 nM of each primer, 3.5 μl of cDNA (cycle: 3 min at 95 °C, 40 cycles 10 s at 95 °C, 30 s at 58 °C). PCR reactions were carried out with a CFX-96 real-time PCR apparatus controlled by an Optical System software (version 3.1) (Bio-Rad Laboratories, CA, USA). All expressions were normalized with housekeeping genes. The sequences of the primers and probes used for the amplifications are given as supplementary data (S8).
CD and FRET experiments
CD spectra have been obtained with a JASCO J-600 spectropolarimeter equipped with a thermostatted cell holder. CD experiments were carried out with oligonucleotides hras-1Y and hras-2Y (3 μM) in 50 mM Tris-acetate, pH from 4.5 to 8, 50 mM KCl. Spectra were recorded in 1 or 0.5 cm quartz cuvette. A thermometer inserted in the cuvette holder allowed a precise measurement of the sample temperature. The spectra have been calculated with J-700 Standard Analysis software (Japan Spectroscopic Co, Ltd) and reported as ellipticity (mdeg) versus wavelength (nm). Each spectrum was recorded three times, smoothed and the baseline subtracted. CD spectra of 3 μM hras-1Y and hras-2Y have been obtained also at various temperatures (20-85 °C), by both heating and cooling the sample solutions (in 50 mM sodium cacodylate pH 5, 50 mM KCl). By plotting the 287 nm ellipticity versus temperature, sigmoidal denaturing and renaturing curves were obtained, which were practically overlapping.
FRET with oligonucleotides hras-1Y and hras-2Y, tagged at the 5′ and 3′ ends with ATTO-488 and TAMRA (as donor we used ATTO-488 because its pH dependence is weaker than that of FAM), were carried out on a Microplate Spectrofluorometer System (Perkin Elmer 2300 Enspire, USA). Each sample contained 50 μl dual-labelled oligonucleotide (200 nM) in 50 mM Tris-acetate buffer, pH from 4.5 to 8, 50 mM KCl and an amount of hnRNP A1 as specified in the figure captions. The samples were incubated at 37 °C as specified in the text. Emission spectra were obtained setting the excitation wavelength at 480 nm and recording the emission from 500 to 650 nm. FRET-melting experiments of hras-1Y and hras-2Y have been performed on a real-time PCR apparatus (CFX96, BioRad, Hercules, CA) in 50 mM sodium cacodylate at pH 5, 5.5, 6, 6.5 and 7, 50 mM KCl. FRET-melting experiments were obtained by increasing the temperature from 20 °C to 95 °C (0.3 °C/min). From the melting data we obtained curves reporting the fraction of folded iM against temperature. These curves were reversible (denaturing and renaturing curves overlapping). The energy transfer (ET) was calculated from the fluorescence intensity of the donor D in the presence (IDA) and absence (ID) of the acceptor as:

IDA and ID were measured in same buffer under identical concentrations (ID was obtained by transforming the dual-labeled oligonucleotide into the corresponding duplex in which the fluorophores are at a distance for which FRET = 0). The FRET efficiency values were converted to distances between donor and acceptor by using:

where R is the distance (Å) and R0 is the Föster distance [defined as the distance at which energy transfer is 50% of the maximum value, assumed to be 50 Å50].
Thermodynamic analysis of reversible iM melting curves
The thermodynamic parameters for the folding of C-rich sequence into the iM conformation were obtained from the melting curves. From ΔG° = −RT ln K = ΔH°–TΔS° (8) it is obtained lnK = (ΔH°/R)(1/T) + ΔS°/R (9). The equilibrium constant K as a function of T is given by K = f/(1–f), where f, the fraction of sequence folded in the iM conformation, is obtained form the melting curves. The plot of ln K versus 1/T gives a straight line whose slope (–ΔH/R) and y-intercept (–ΔS/R) provide the thermodynamic parameters (S9).
PAGE assays
Oligonucleotides hras-1Y and hras-2Y were end-labelled with [γ-32P]ATP and T4 polynucleotide kinase. For competition experiments we used a DNA chemically labelled to dy-781. Before EMSA, the iM-forming oligonucleotides were allowed to form their structure in 50 mM Tris–acetate, pH 5.5, 50 mM KCl, (overnight incubation at room temperature). Radiolabelled oligonucleotides (10 nM) were incubated for 30 min at 20 °C with increasing amounts of hnRNP A1 (0–12 μg) as specified in Fig. 4B, in 50 mM Tris–acetate, pH 5.5, 50 mM KCl, 1 mM DTT, 8% glycerol, 1% Phosphatase Inhibitor Cocktail I (Sigma, Milan, Italy), 5 mM NaF, 1 mM Na3VO4, 2.5 ng/μl salmon sperm DNA (binding buffer). After incubation, the reaction mixtures were loaded in 5% PAGE in 50 mM Tris-acetate pH 5.5, thermostatted at 20 °C. After running the gel was dried and exposed to autoradiography (G E Healthcare, Milan) for 16 h at –80 °C. Mobility-shift experiments of cold hras-1Y and hras-2Y have been performed on 15% PAGE, 25 mM KCl, at pH 5 (50 mM sodium acetate) or pH 7 (50 mM Tris-acetate), 20 °C. 20% PAGE in denaturing 7 M urea conditions, was carried out in TBE. The gels were stained with “stains-all” dye. Competition assay with 28 nM hras-1Y-dy781 were performed at 37 °C with 3 μM of hnRNP A1 (100-fold over iM hras-1Y-dy781) and competitor oligonucleotides (150-fold over iM) in 50 mM Tris-acetate, pH 5.5, 50 mM KCl, 1 mM EDTA, 2.5ng/μl Salmon sperm. After incubation, the reaction mixtures were loaded in 5% PAGE 1xTBE, thermostated at 20 °C. After running the gel was analysed by Odyssey CLx scanner /ImageStudio Software (Li-Cor Biosciences).
Cell culture and transfections
T24 human urinary bladder cancer cells were maintained in exponential growth in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 100 U/ml penicillin, 100 mg/ml streptomycin, 20 mM L-glutamine and 10% fetal bovine serum (Euroclone, Milan, Italy).
For transfection we plated 10000 cells for each well in a 96 well plate and transfected using Jet PEI (Polyplus Illkirch FRANCE) following manufacturers in vitro protocol for DNA oligonucleotides transfection with 400 nM oligonucleotide (48 pmol) and N/P = 3.
Additional Information
How to cite this article: Miglietta, G. et al. GC-elements controlling HRAS transcription form i-motif structures unfolded by heterogeneous ribonucleoprotein particle A1. Sci. Rep. 5, 18097; doi: 10.1038/srep18097 (2015).
References
Lowy, D. R. & Willumsen, B. M. Function and regulation of ras. Annu. Rev. Biochem. 62, 851–891 (1993).
Porter, A. C. & Vaillancourt, R. R. Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene 17, 13434–13452 (1998).
Cogoi, S., Shchekotikhin, A. E. & Xodo, L. E. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 42, 8379–8388 (2014).
Membrino, A., Cogoi, S., Pedersen, E. B. & Xodo, L. E. G4-DNA formation in the HRAS promoter and rational design of decoy oligonucleotides for cancer therapy. PLoS One 6, e24421 (2011).
Kendrick, S., Akiyama, Y., Hecht S. M. & Hurley, L. H. The i-motif in the bcl-2 P1 promoter forms an unexpectedly stable structure with a unique 8:5:7 loop folding pattern. J. Am. Chem. Soc. 131, 17667- 17676 (2009).
Kendrick, S. et al. The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 136, 4161–4171 (2014).
Kang, H. J., Kendrick, S., Hecht, S. M. & Hurley, L. H. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 136, 4172–4185 (2014).
Cui, Y. et al. Molecular population dynamics of DNA structures in a bcl-2 promoter sequence is regulated by small molecules and the transcription factor hnRNP LL. Nucleic Acids Res. 42, 5755–5764 (2014).
Gehring, K., Leroy, J. L. & Guéron, M. A. tetrameric DNA structure with protonated cytosine-cytosine base pairs. Nature 363, 561–565 (1993).
Manzini, G., Yathindra N. & Xodo, L. E. Evidence for intramolecularly folded i-DNA structures in biologically relevant CCC-repeat sequences. Nucleic Acids Res. 22, 4634–4640 (1994).
Day, A. H., Pavlou, P. & Waller, Z. A. E. i-Motif DNA: structure, stability and targeting with ligands. Bioorg. & Med. Chem. 22, 4407–4418 (2014).
Mergny, J. L., Lacroix, L., Han, X., Leroy, J. L. & Hélène, C. Intramolecular folding of pyrimidine oligonucleotides into an i-DNA motif. J. Am. Chem. Soc. 117, 8887–8898 (1995).
Mathur, V., Verma, A., Maiti, S. & Chowdhury S. Thermodynamics of i-tetraplex formation in the nuclease hypersensitive element of human c-myc promoter. Bioch. Biophys. Res. Commun. 320, 1220–1227 (2004).
Kaushik, M., Suehl, N. & Marky, L. A. Calorimetric unfolding of the biomolecular and i-motif complexes of the human telomere complementary strand, d(C3TA2)4. Bioph. Chemistry 126, 154–164 (2007).
Gargallo, R. Hard/soft hybrid modeling of temperature-induced unfolding processes involving G-quadruplex and i-motif nucleic acid structures. Analytical Biochem. 466, 4–15 (2014).
Bhavsar-jog, Y. P., Dornshuld, E., van Brooks, T. A., Tschumper, G. S. & Wadkins, R. M. Epigenetic modification, dehydration and molecular crowding effects on the thermodynamics of i-motif structure formation from C-rich DNA. Biochemistry 53, 1586–1594 (2014).
Cogoi, S., Paramasivam, M., Spolaore, B. & Xodo, L. E. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 36, 3765–3780 (2008).
Cogoi, S. & Xodo, L. E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 34, 2536–2549 (2006).
Rankin, S. et al. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 127, 10584–10589 (2005).
Fernando, H. et al. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry 45, 7854–7860 (2006).
Siddiqui-Jain, A., Grand, C. L., Bearss, D. J. & Hurley, L. H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 99, 11593–11598 (2002).
Hurley, L. H., Von Hoff, D. D., Siddiqui-Jain, A. & Yang, D. Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element. Semin Oncol. 33, 498–512 (2006).
Balasubramanian, S., Hurley, L. H. & Neidle, S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug. Discov. 10, 261–275 (2011).
Brooks, T. A. & Hurley, L. H. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. Cancer 9, 849–861 (2009).
Bucek, P., Jaumot, J., Aviñó, A., Eritja, R. & Gargallo, R. pH-Modulated Watson–Crick Duplex–Quadruplex Equilibria of Guanine-Rich and Cytosine-Rich DNA Sequences 140 Base Pairs Upstream of the c-kit Transcription Initiation Site. Chemistry 15, 12663–12671 (2009).
Xu, Y. & Sugiyama, H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb). Nucleic Acids Res. 34, 949–954 (2006).
Guo, K. et al. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 129, 10220–10228 (2007).
Sun, D. & Hurley, L. H. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 52, 2863–74 (2009).
Selvam, S., Koirala, D., Yu, Z. & Mao, H. Quantification of topological coupling between DNA superhelicity and G-quadruplex formation. J. Am. Chem. Soc. 136, 13967–13970 (2014).
Miyoshi, D., Matsumura, S., Nakano, S. & Sugimoto, N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 126, 165–169 (2004).
Zhao, C., Ren, J. & Qu, X. Single-walled carbon nanotubes binding to human telomeric i-motif DNA under molecular-crowding conditions: more water molecules released. Chemistry 14, 5435–5439 (2008).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Saenger, W. Principles of Nucleic Acid Structures. (Ed. Springer-Verlag, New York 1984).
Chen, C. Study of pH-induced folding and unfolding kinetics of the DNA i-motif by stopped-flow circular dichroism. Langmuir 28, 17743–17748 (2012).
Dhakal, S. et al. Coexistence of an ILPR i-motif and a partially folded structure with comparable mechanical stability revealed at the single-molecule level. J. Am. Chem. Soc. 132, 8991–8997 (2010).
Smiatek, J., Chen, C., Liu, D. & Heuer, A. J. Stable conformations of a single stranded deprotonated DNA i-motif. J. Phys. Chem. B. 115, 13788–13795 (2011).
Yang, B. & Rogers, M. T. Base-pairing energies of proton-bound heretodimers of cytosine and modified cytosines: implications for the stability of DNA i-motif conformations. J. Am. Chem. Soc. 136, 282–288 (2014).
Michelotti, E. F., Michelotti, G. A., Aronsohn, A. I. & Levens, D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 16, 2350–2360 (1996).
Takimoto, M. et al. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem. 268, 18249–18258 (1993).
Jean-Philippe, J., Paz, S. & Caputi, M. hnRNP A1: the Swiss army knife of gene expression. Int. J. Mol. Sci. 14, 18999–19024 (2013).
Ushigome, M. et al. Up-regulation of hnRNP A1 gene in sporadic human colorectal cancers. Int. J. Oncol. 26, 635–640 (2005).
Ding, J. et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 13, 1102–1115 (1999).
Campbell, M. A. & Wengel, J. Locked vs. unlocked nucleic acids (LNA vs. UNA): contrasting structures work towards common therapeutic goals. Chem. Soc. Rev. 40, 5680–5689 (2011).
Pasternak, A. & Wengel, J. Unlocked nucleic acid–an RNA modification with broad potential. Org. Biomol. Chem. 9, 3591–3597 (2011).
Brunner, S., Fürtbauer, E., Sauer, T., Kursa, M. & Wagner, E. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear PEI or electroporation. Mol. Ther. 5, 80–86 (2002).
Fukuda, H. et al. Unfolding of quadruplex structure in the G-rich strand of the minisatellite repeat by the binding protein UP1. Proc. Natl. Acad. Sci. USA 99, 12685–12690 (2002).
Dhakal, S. et al. G-quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA. Biophys J. 102, 2575–2584 (2012).
Paramasivam, M. et al. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 37, 2841–2853 (2009).
Clegg, R. M. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 211, 353–388 (1990).
Acknowledgements
This work was supported by AIRC (The Italian Association for Cancer Research; IG2013, Project Code 14301). We thank Dolores Ross for proofreading the manuscript.
Author information
Authors and Affiliations
Contributions
L.X. and S.C. conceived the study, G.M. and S.C. performed the experiments, E.P. designed and synthesized the decoys, L.X. wrote the main manuscript text, all authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Miglietta, G., Cogoi, S., Pedersen, E. et al. GC-elements controlling HRAS transcription form i-motif structures unfolded by heterogeneous ribonucleoprotein particle A1. Sci Rep 5, 18097 (2016). https://doi.org/10.1038/srep18097
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep18097
This article is cited by
-
Theranostic approach to specifically targeting the interloop region of BCL2 i-motif DNA by crystal violet
Scientific Reports (2023)
-
Secondary structural choice of DNA and RNA associated with CGG/CCG trinucleotide repeat expansion rationalizes the RNA misprocessing in FXTAS
Scientific Reports (2021)
-
Preferential targeting cancer-related i-motif DNAs by the plant flavonol fisetin for theranostics applications
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.