Abstract
Over-expression of de novo lipogenesis (DNL) pathway genes is associated with the prognosis of various types of cancers. However, effects of single nucleotide polymorphisms (SNPs) in these genes on recurrence and death of hepatocellular carcinoma (HCC) patients after surgery are still unknown. A total of 492 primary HCC patients treated with surgery were included in this study. Nine SNPs in 3 genes (ACACA, FASN and ACLY) of DNL pathway were genotyped. Multivariate Cox proportional hazard regression model and Kaplan-Meier curve were used to analyze the association of SNPs with clinical outcomes. Two SNPs in ACACA gene were significantly associated with overall survival of HCC patients. Patients carrying homozygous variant genotype (VV) in rs7211875 had significantly increased risk of death, while patients carrying VV genotype in rs11871275 had significant decreased risk of death, when compared with those carrying homozygous wild-type or heterozygous genotypes. Moreover, patients carrying VV genotype in rs11871275 had decreased recurrence risk, while patients carrying variant genotype in rs4485435 of FASN gene had increased recurrence risk. Further cumulative effect analysis showed significant dose-dependent effects of unfavorable SNPs on both death and recurrence. SNPs in DNL genes may serve as independent prognostic markers for HCC patients after surgery.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common solid tumor and the fourth leading cause of cancer-related death worldwide, especially in East Asia. Even with aggressive treatment, HCC usually has a poor prognosis, with a 5-year survival rate as low as 25%–39% after standard treatment1. Traditional clinicopathological parameters such as imaging parameters, serum alpha fetoprotein level and tumor stage only offer limited efficacy for prognosis prediction and fail to effectively guide the individualized therapeutics for HCC2. Therefore, it is extremely urgent to develop novel biomarkers to discriminate patient groups with different clinical outcomes and improve the treatment for HCC patients.
One of the important features of cancer cells is the increased de novo lipogenesis (DNL) for the purpose to meet the excessive bioenergetic and structural demands of proliferation, irrespective of the extracellular lipid level3. The newly synthesized lipids have also been demonstrated to mediate cell migration, signal transduction, intracellular trafficking by constructing membrane rafts4,5,6. Thus, the enhanced de novo lipogenesis (DNL) plays important roles in tumor cell survival7,8,9,10. At the molecular level, enhanced DNL is reflected by the coordinately increased activity or expression of lipogenic enzymes in neoplastic cells3. There are three key rate-limiting enzymes involved in DNL pathway, i.e. ATP citrate lyase (ACLY), acetyl-CoA carboxylase alpha (ACACA) and fatty-acid synthase (FASN). ACLY converts citrate into cytosolic acetyl-CoA, which is transformed into malonyl-CoA by the catalysis of ACACA. The malonyl-CoA product is further converted by FASN to long-chain fatty acids. Numerous studies have demonstrated that DNL enzymes are overexpressed in various human cancer types, including liver cancer11,12. Furthermore, the overexpression of FASN or ACACA in tumor tissues has been associated with poor clinical outcomes of prostate, breast, nasopharyngeal, colon, ovarian and lung cancer patients5,13,14,15,16,17,18. ACLY is upregulated in several types of cancer cells and this enzyme also plays an important role in cancer cell progression19. Therefore, molecules in DNL pathway may have potentials to serve as prognostic markers for cancer patients.
Single nucleotide polymorphism (SNP), a common typical genomic variation, can be used as a stable biomarker of genetic background to profile and predict some useful characteristics about cancer, including the susceptibility, development of cancer and patients' prognosis and response to treatment. Also, genetic variations may be important cause factors that influence the expression or catalytic activity of DNL enzymes and subsequently modulate their biological functions20. Quite a few studies have investigated the relationship between single nucleotide polymorphisms (SNPs) of DNL genes and development or prognosis of several tumors such as uterine leiomyomata, breast and prostate cancers21,22,23,24. However, no studies have focused on the association between polymorphisms of DNL genes and HCC prognosis until now. Therefore, we assessed the effects of nine SNPs in the three key genes of DNL pathway on the prognosis of HCC patients in a cohort of 492 Chinese subjects.
Methods
Study Population
Between January 2009 and January 2012, a total of 518 HCC patients were enrolled at Xijing Hospital affiliated with Fourth Military Medical University in Xi'an and the Eastern Hepatobiliary Surgery Hospital affiliated with Secondary Military Medical University in Shanghai, China. Patients with histologically confirmed primary HCC were eligible for the enrollment. There were no recruitment restrictions on age, gender and tumor stage. All HCC diagnosis was based on the National Comprehensive Cancer Network (NCCN) clinical practice guidelines in oncology. In the present study, we excluded 26 patients, including 3 patients with incomplete clinical data and 23 patients who were dead within 1 month post-surgery. Finally, a total of 492 eligible HCC patients were recruited, with complete and validated demographic, clinical and follow-up data. All patients received surgery within 2 months after diagnosis. The primary tumor was completely resected for all patients and was confirmed by pathological review of the tumors after resection. No patient received anticancer treatment before surgery. The study was approved by the Ethical Committees of Fourth Military Medical University and Secondary Military Medical University. Written consent was obtained from each patient. All experiments involving human subjects and tissues were performed in accordance with guidelines approved by Fourth Military Medical University and Secondary Military Medical University.
Epidemiologic and clinical data collection
Demographic data were collected through in-person interviews at the time of initial visit or follow-up in the clinics, medical chart review, or consultation with the treating physicians by trained clinical research specialists. For data acquired from multiple sources, the research staff compared and validated the consistency of these data. The patients and/or family members were further contacted for verification if there were discrepancies. Disease stage was assessed using the seventh American Joint Committee on Cancer (AJCC) and Barcelona Clinic Liver Cancer (BCLC) staging systems. The follow-up information was updated at 6-month intervals through onsite interviews, direct calling, or medical chart review. The latest follow-up in this study was carried out in January 2013.
SNP selection and genotyping
Genomic DNA was obtained for each participant as described previously25. Three DNL pathway genes (ACACA, FASN and ACLY) were included as candidate genes in this study to estimate the effect of their genetic polymorphisms on the survival in HCC patients. SNPs in these genes were selected using a set of web-based SNP selection tools (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm) as previously described26. Briefly, the strategy and criteria for the selection of SNPs were as follows: a) located in target genes or their 2000 bp flanking regions; b) potentially functional SNPs had priority; c) had minor allele frequency (MAF) >5% in the Asian population (CHB). If there were multiple potentially functional SNPs within the same block (defined by the linkage coefficient r2 > 0.80), only 1 SNP was included. Functional SNPs included missense SNPs in exons and SNPs in microRNA binding sites of 3′ untranslated region, transcription factor binding site of the 5′ flanking region and splice sites.
Through the selection process, 9 SNPs were selected for prognosis analysis in this study, including 3 SNPs in transcription factor binding site sequences (rs11871275, rs9912300 and rs11653012), 2 missense SNPs (rs7211875 and rs2304497), 2 synonymous SNPs (rs1140616 and rs4485435); 1 splice site SNP (rs1714987) and 1 SNP with unknown function (rs4246444 PMID: 21646813) (Table S1). Genotyping was performed with iPLEX (Sequenom, San Diego, CA) matrix-assisted laser desorption/ionization time-of-flight mass spectrometry technology. Laboratory personnel conducting genotyping were blinded to patient information. Strict quality control measures were implemented during genotyping with more than 99% concordance in samples that were randomly selected to be genotyped in duplicate.
Immunohistochemical staining of ACACA and FASN
Formalin-fixed, paraffin-embedded HCC tissues from 64 patients (including 57 males and 7 females) were collected. The median age of these patients were 54 years (range 18–78). There were 52 cases were diagnosed with TNM stage I or II diseases, while 20 with well- or moderately-differentiated tumors. H&E slides from these patients were viewed under a light microscope by a pathologist and 4-μm-thick tissue sections were cut from corresponding blocks containing representative tumor regions. After deparaffinization with dimethylbenzene, the tissue sections were rehydrated through 100%, 95%, 90%, 85% and 75% ethanol. After three washes in phosphate-buffered saline (PBS), every five minutes, the slides were boiled in antigen retrieval buffer containing 0.01 mol/L citrate antigen retrieval (pH = 6.0) in the pressure cooker and then rinsed in peroxidase quenching solution (Invitrogen) to block endogenous peroxidase. The sections were then incubated with a rabbit anti-human ACACA antibody (1:200, Abcam) or rabbit anti-human FASN antibody (1:200, Abcam) at 4°C overnight and then with an Broad Spectrum Second Antibody (Invitrogen) at 37°C for 20 min. After three washes, the visualization signal was developed with Invitrogen Histostain Plus kit. The intensity and extent of immunostaining were evaluated for all samples under double-blinded conditions. In brief, the percentage of positive staining was scored as 0 (0–9%), 1 (10%–25%), 2 (26%–50%), 3 (51%–75%) or 4 (76%–100%) and the intensity as 0 (no staining), 1 (weak staining), 2 (moderate staining) or 3 (dark staining). The total score was calculated as the product of intensity and extent, ranging from 0 to 12.
Statistical analysis
All statistical analyses were performed using the SPSS Statistics 19.0 software (IBM). The 3 genetic models (additive, dominant and recessive) were applied to assess the association of single SNPs with clinical outcome of HCC patients. The best-fitting model was defined as that with the smallest P value. Only the result predicted by the best model was reported and considered in the subsequent analysis. Hazard ratio (HR) was estimated in a multivariate Cox proportional hazard model, adjusting for age, gender, TNM stage, tumor differentiation and treatment regimen. The cumulative effect of unfavorable genotypes on OS or RFS was estimated in Cox model27. Adjusted Kaplan–Meier curve and log-rank test were used to assess the differences in overall and recurrence-free survival (OS and RFS respectively) between patients with different genotypes28. All statistical analyses were two-sided and P < 0.05 was considered to be of statistical significance.
Results
Clinical characteristics and their effects on prognosis of HCC patients after surgery
A total of 492 HCC patients were included in this study, with a median age of 53 years (ranging from 18 to 79 years). More than 90% of patients (n = 446) were positive for serum HBsAg. The numbers of patients in different TNM stages were as follows: 394 for stage I and II (80.1%), 98 for stage III and IV (19.9%). The majority (n = 344, 70.1%) of patients had poor differentiated or undifferentiated tumors, while 57.5% (n = 283) had a tumor size larger than 5.0 cm and 19.5% (n = 96) had multiple lesions. More than 70% (n = 362) of patients were at BCLC stage A and 37.6% (n = 185) received adjuvant transcatheter arterial chemoembolization treatment (TACE).
During the median follow-up of 21.8 months (ranging from 1.6 to 48.3 months), 188 (38.2%) patients died of HCC and 309 developed recurrence. Multivariate Cox regression analysis showed that serum alpha fetoprotein (AFP) level, tumor differentiation, TNM stage, BCLC stage and PVTT had significant influence on overall survival (OS) and recurrence-free survival (RFS) for HCC patients (P < 0.01 for all). Adjuvant TACE therapy has been shown to reduce risks of both death (HR, 0.62; 95% CI, 0.45–0.85; P < 0.01) and recurrence (HR, 0.56; 95% CI 0.43–0.71; P < 0.01) (Table 1).
Association of single SNP with clinical outcomes of HCC patients
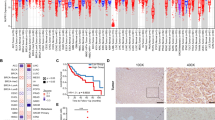
We assessed the effects of 9 SNPs in three DNL pathway genes (ACACA, FASN and ACLY) on the death and recurrence in HCC patients using the multivariate cox regression model (Table 2). After adjusting for gender, age, HBsAg status, AFP level, TNM stage and differentiation, our data showed that SNPs rs7211875 and rs11871275 in ACACA gene were significantly associated with the death risk of HCC patients under recessive model (for rs7211875, HR = 2.13, 95% CI = 1.17–3.88; P = 0.01; for rs11871275, HR = 0.22, 95% CI = 0.06–0.91; P = 0.04). Adjusted survival curve showed similar results, indicating that HCC patients carrying the homozygous variant (CC) genotype in rs7211875 had significantly worse OS than those carrying the other genotypes (P = 0.01, Figure 1A), whereas patients with homozygous variant (TT) genotype in rs11871275 had better OS than those with other genotypes (P = 0.04, Figure 1B). Moreover, our data showed that TT genotype in rs11871275 of ACACA gene reduced the recurrence risk of HCC when compared to patients with AA and AT genotypes (HR = 0.41, 95% CI = 0.17–1.00; P = 0.05), whereas variant genotype (GC/CC) in rs4485435 of FASN gene was associated with increased recurrence risk, when compared to patients with GG genotype (HR = 1.32; 95% CI = 1.02–1.72, P = 0.03). Kaplan-Meier analysis confirmed these findings, showing that HCC patients carrying the TT genotype in rs11871275 had better OS than those carrying the other genotypes (P = 0.05, Figure 1C), while patients with GC/CC genotypes in rs4485435 trended to show worse OS than did those with GG genotype (P = 0.09, Figure 1D).
Survival curves of OS (A and B) and RFS (C and D) of HCC patients stratified by SNPs.
Homozygous wild-type genotype, heterozygous genotype and homozygous variant genotype for rs7211875 was TT, TC, CC, respectively; for rs11871275 was AA, AT, TT, respectively; for rs4485435 was GG, GC, CC, respectively.
Cumulative effects of unfavorable genotypes on the prognosis of HCC patients
In order to assess the cumulative effects of genetic variants on HCC patients' outcomes, we did a joint analysis by including the three unfavorable genotypes determined by the single SNP analysis. Briefly, homozygous variant genotype (CC) for rs7211875, homozygous wild-type and heterozygous genotype (AA+AT) for rs11871275 and homozygous wild-type and heterozygous genotype (GC+CC) for rs4485435 were defined as the unfavorable genotypes. When using group 1 (with 0 unfavorable genotype) as reference, HCC patients in group 3 (with 2 unfavorable genotypes) had a 6.3-fold increased risk of death (95% CI, 1.41–28.16; P < 0.01) (Table 3). Furthermore, when compared with group 1 with 0 unfavorable genotype, increased recurrence risk was observed in group 2 with 1 unfavorable genotype (HR, 2.72; 95% CI, 1.00–7.34; P = 0.05) and group 3 with 2 unfavorable genotypes (HR, 3.43; 95% CI, 1.25–9.43; P = 0.02) (Table 4). The cumulative death or recurrence risks of unfavorable genotypes exhibited significant dose-dependent manners with both P for trend <0.01. Similarly, Kaplan-Meier curves showed significant different OS and RFS among HCC patient groups stratified by number of unfavorable genotypes (P = 0.05 and P = 0.01, respectively, Figure 2).
The cumulative effects of unfavorable genotypes in ACACA and FASN genes on overall (A) and recurrence-free (B) survival of HCC patients analyzed by Kaplan-Meier curves.
Group1, patients with 0 unfavorable genotype; Group2, patients with 1 unfavorable genotype; Group3, patients with 2 unfavorable genotypes.
Relationship between SNP genotypes and the expression of ACACA and FASN
We have examined the expression of ACACA and FASN in 64 HCC tissue samples using immunohistochemistry. As showed in supplementary figure 1, patients carrying AT or AA genotype of rs11871275 exhibited significantly higher expression of ACACA when compared with those carrying TT genotype, while the genotypes of SNP rs7211875 and rs4485435 did not show significant association with the expression of FASN. Due to the limited sample size, our results were very preliminary and need to be validated in larger cohort.
Discussion
In the present study, we evaluated the effects of 9 SNPs in three DNL pathway genes (ACACA, FASN and ACLY) on the prognosis of Chinese HCC patients with surgical resection. Our data demonstrated that SNPs rs7211875 and rs11871275 in the ACACA gene were significantly associated with the OS of HCC patients, whereas rs11871275 in ACACA gene and rs4485435 in FASN gene were significantly associated with the RFS of patients. Furthermore, we found an accumulative risk of both death and recurrence in HCC patients with increasing number of unfavorable genotypes. To the best of our knowledge, this is the first study to report significant effects of genetic variants in DNL pathway genes on the clinical outcomes of HCC patients.
It has been well recognized that the up-regulation of the fatty acid biosynthetic activity starts at a relatively early stage in various types of tumors29,30. Cancer cells seem to be highly dependent on DNL for their proliferation and survival29. Recently, quite a few studies have demonstrated that SNPs in DNL pathway genes are associated with the risk and prognosis of several tumors21,22,23,24. For example, it has been reported that SNPs in the FASN gene are associated with the risk of uterine leiomyomata21, while SNPs in the ACACA gene are associated with the risk of breast cancer22,23. Nguyen PL et al. have demonstrated that SNPs in FASN gene are associated with the risk and prognosis of prostate cancer24. However, no studies have focused on the association between polymorphisms of DNL pathway genes and HCC prognosis until now.
In the present study, we found that two SNPs (rs7211875 and rs11871275) in the ACACA gene were significantly associated with the risk of death of HCC patients, while SNP rs11871275 in ACACA and rs4485435 in FASN gene were associated with the RFS of HCC patients. Our findings are supported by previous studies. Calvisi DF et al. has reported that induction of DNL pathway, an important effector pathway of the oncogenic AKT/mTORC1 axis, has pathogenic and prognostic significance for HCC31. Moreover, elevated FASN expression is associated with the progression of HCC and poor prognosis of patients32. Compared with FASN, little is known about the biological functions and clinical significance of ACACA in HCC. However, marked elevation of ACACA expression and activity has been reported in breast cancer cells33,34. In addition, ACACA expression is essential to promote breast cancer cell survival10. Since rate-limiting catalysis of ACACA in fatty acid elongation in DNL pathway, it is rational to hypothesis that ACACA may also play an important role in the development of HCC.
The biological function of the three SNPs and the molecular mechanisms through which they affect gene functions and clinical outcome of HCC remain to be elucidated. Silicon analysis (including more than 80 bp upstream and downstream of alternative site) with University of California Santa Cruz genome browser (http://genome.ucsc.edu/) indicated that rs4485435 in the exon 21 of FASN may be involved in the alternative splicing of FASN mRNA. Transcription factor binding sites (TFBS) analysis demonstrated that three SNPs, rs7211875, rs11871275 and rs4485435 are close to one or two TFBS sites, indicating their potential impact on the transcription of DNL genes. MultiZ's alignment35 estimated that none of the three SNPs are located in the highly conserved regions of the genome. Our preliminary immunohistochemical analysis showed that that AT+AA genotypes of rs11871275 were significantly associated with higher expression level of ACACA when compared to TT genotype, which is in line with notion that elevated ACACA expression promotes the proliferation of cancer cells. However, no significant association of other SNPs with the expression level of either ACACA or FASN, suggesting that these SNPs may affect the biological activity but not the expression of these proteins. Therefore, these findings need to be validated in larger patient cohort and their biological functions need to be further investigated. Moreover, we did not observe any significant association between 2 functional SNPs in ACLY gene and the prognosis of HCC patients. Possible explanation for our results is that these two SNPs may not or just very weakly affect the expression and functions of ALCY in HCC, which need further investigation in future study.
Various cellular pathways and a complex molecular network may play key roles in the development of HCC. Given the limited power of single SNP in predicting progression of a given disease, we applied a pathway-based approach in our study to evaluate the biological interplay between genes. Our data provide insights into the way by which multiple genes contribute to the pathogenesis of HCC. Furthermore, in this study, we also did a joint analysis by including the three unfavorable genotypes and found a significant dose-dependent effect of the unfavorable genotypes in DNL gene on the OS and RFS. Our results indicate that more powerful prognostic or predictive value can be obtained based on the combination of single SNPs. Therefore, the combination of patients' clinicopathological parameters and genetic variants status of DNL pathway will promote the accuracy and sensitivity of prediction and help improve the clinical management for HCC patients.
Our study has several strengths. First, our pathway-based approach is a logical extension of the candidate gene method, avoiding the requirement of a much larger sample size for a genome-wide association study with free-hypothesis method. Second, the patients are recruited from two large liver disease clinics in China, which can help to minimize the potential selection bias. Our study also has limitations. First, our sample size may not be large enough to detect minimal associations and interactions. Second, we recognize that our follow-up time is relatively short (median follow-up time, 21.8 months; range, 1.6–48.3 months). However, the disadvantage may not affect our results too much because our population has already presented with a considerable percentage of deaths and recurrence.
Conclusions
Overall, as the first study observing the impact of DNL gene variants on HCC prognosis, our data strongly suggest that SNPs in ACACA and FASN genes may serve as independent prognostic markers for OS and RFS prediction in HCC patients after surgery treatment. These findings warrant validation in other patient populations and further studies on the mechanism underlying the effects of these SNPs on prognosis in HCC.
References
Thomas, M. B. & Zhu, A. X. Hepatocellular carcinoma: the need for progress. J Clin Oncol. 23, 2892–9 (2005).
Mann, C. D. et al. Prognostic molecular markers in hepatocellular carcinoma: a systematic review. Eur J Cancer. 43, 979–992 (2007).
Swinnen, J. V., Brusselmans, K. & Verhoeven, G. Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care. 9, 358–65 (2006).
Swinnen, J. V. et al. Androgens, lipogenesis and prostate cancer. J Steroid Biochem Mol Biol. 92, 273–9 (2004).
Shurbaji, M. S., Kalbfleisch, J. H. & Thurmond, T. S. Immunohistochemical detection of a fatty acid synthase (OA-519) as a predictor of progression of prostate cancer. Hum Pathol. 27, 917–21 (1996).
Simons, K. & Toomre, D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 1, 31–9 (2000).
Li, N. et al. Fatty acid synthase regulates proliferation and migration of colorectal cancer cells via HER2-PI3K/Akt signaling pathway. Nutr Cancer. 64, 864–70 (2012).
Migita, T. et al. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst. 101, 519–32 (2009).
Lin, R. et al. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell. 51, 506–18 (2013).
Chajes, V., Cambot, M., Moreau, K., Lenoir, G. M. & Joulin, V. Acetyl-CoA carboxylase alpha is essential to breast cancer cell survival. Cancer Res. 66, 5287–94 (2006).
Kuhajda, F. P. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 16, 202–8 (2000).
Calvisi, D. F. et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 140, 1071–1083. e5 (2011).
Alo, P. L. et al. Expression of fatty acid synthase (FAS) as a predictor of recurrence in stage I breast carcinoma patients. Cancer. 77, 474–82 (1996).
Kao, Y. C. et al. Fatty acid synthase overexpression confers an independent prognosticator and associates with radiation resistance in nasopharyngeal carcinoma. Tumour Biol 34, 759–68 (2013).
Ogino, S. et al. Cohort study of fatty acid synthase expression and patient survival in colon cancer. J Clin Oncol. 26, 5713–20 (2008).
Rahman, M. T. et al. Fatty acid synthase expression associated with NAC1 is a potential therapeutic target in ovarian clear cell carcinomas. Br J Cancer. 107, 300–7 (2012).
Cerne, D., Zitnik, I. P. & Sok, M. Increased fatty acid synthase activity in non-small cell lung cancer tissue is a weaker predictor of shorter patient survival than increased lipoprotein lipase activity. Arch Med Res. 41, 405–9 (2010).
Wang, Y. et al. Prognostic and therapeutic implications of increased ATP citrate lyase expression in human epithelial ovarian cancer. Oncol Rep. 27, 1156–62 (2012).
Zaidi, N., Swinnen, J. V. & Smans, K. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res. 72, 3709–14 (2012).
Laing, R. E., Hess, P., Shen, Y., Wang, J. & Hu, S. X. The role and impact of SNPs in pharmacogenomics and personalized medicine. Curr Drug Metab. 12, 460–86 (2011).
Eggert, S. L. et al. Genome-wide linkage and association analyses implicate FASN in predisposition to Uterine Leiomyomata. Am J Hum Genet. 91, 621–8 (2012).
Sinilnikova, O. M. et al. Haplotype-based analysis of common variation in the acetyl-coA carboxylase alpha gene and breast cancer risk: a case-control study nested within the European Prospective Investigation into Cancer and Nutrition. Cancer Epidemiol Biomarkers Prev. 16, 409–15 (2007).
Sinilnikova, O. M. et al. Acetyl-CoA carboxylase alpha gene and breast cancer susceptibility. Carcinogenesis. 25, 2417–24 (2004).
Nguyen, P. L. et al. Fatty acid synthase polymorphisms, tumor expression, body mass index, prostate cancer risk and survival. J Clin Oncol. 28, 3958–64 (2010).
Xing, J. et al. GWAS-identified colorectal cancer susceptibility locus associates with disease prognosis. Eur J Cancer. (Oxford, England: 1990) 47, 1699–1707 (2011).
Zhou, F. et al. Functional polymorphisms of circadian positive feedback regulation genes and clinical outcome of Chinese patients with resected colorectal cancer. Cancer. 118, 937–46 (2012).
Cox, D. R. Regression models and life-tables. J R Stat Soc A Stat. Series B (Methodological), 34, 187–220 (1972).
Kaplan, E. L. & Meier, P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 53, 457–481 (1958).
Menendez, J. A. & Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 7, 763–777 (2007).
Piyathilake, C. J. et al. The expression of fatty acid synthase (FASE) is an early event in the development and progression of squamous cell carcinoma of the lung. Hum Pathol. 31, 1068–1073 (2000).
Calvisi, D. F. et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 140, 1071–83 (2011).
Zhu, X. et al. Combined Phosphatase and Tensin Homolog (PTEN) Loss and Fatty Acid Synthase (FAS) Overexpression Worsens the Prognosis of Chinese Patients with Hepatocellular Carcinoma. Int J Mol Sci. 13, 9980–91 (2012).
Yoon, S. et al. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. J Biol Chem. 282, 26122–31 (2007).
Milgraum, L. Z., Witters, L. A., Pasternack, G. R. & Kuhajda, F. P. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin Cancer Res. 3, 2115–20 (1997).
Blanchette, M. et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14, 708–715 (2004).
Acknowledgements
This work was supported by Program for New Century Excellent Talents in University, National Basic Research Program of China (No.2009CB521704), National Natural Science Foundation of China (No.81302054 and 81272522) and National Key Scientific and Technological Project (No.2011ZX09307-001-04).
Author information
Authors and Affiliations
Contributions
Q.H. and K.Z. conceived and designed the experiments. H.J. and J.D. performed the experiments. P.Q., J.L. and Y. C. analyzed the data. Y.Y., J.H. and C.Y. contributed sample collection. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
supplementary figure 1 and table 1
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Jiang, H., Dai, J., huang, X. et al. Genetic variants in de novo lipogenic pathway genes predict the prognosis of surgically-treated hepatocellular carcinoma. Sci Rep 5, 9536 (2015). https://doi.org/10.1038/srep09536
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep09536
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.