Abstract
MPIase is a glycolipid that is involved in membrane protein integration. Despite evaluation of its functions in vitro, the lack of information on MPIase biosynthesis hampered verification of its involvement in vivo. In this study, we found that depletion of CdsA, a CDP-diacylglycerol synthase, caused not only a defect in phospholipid biosynthesis but also MPIase depletion with accumulation of the precursors of both membrane protein M13 coat protein and secretory protein OmpA. Yeast Tam41p, a mitochondrial CDP-diacylglycerol synthase, suppressed the defect in phospholipid biosynthesis, but restored neither MPIase biosynthesis, precursor processing, nor cell growth, indicating that MPIase is essential for membrane protein integration and therefore for cell growth. Consistently, we observed a severe defect in protein integration into MPIase-depleted membrane vesicles in vitro. Thus, the function of MPIase as a factor involved in protein integration was proven in vivo as well as in vitro. Moreover, Cds1p, a eukaryotic CdsA homologue, showed a potential for MPIase biosynthesis. From these results, we speculate the presence of a eukaryotic MPIase homologue.
Similar content being viewed by others
Introduction
Protein integration into biological membranes is a vital process in all organisms. The molecular mechanisms underlying protein integration are widely conserved, as the main components involved in protein integration, such as SRP (signal recognition particle), SR (SRP receptor), SecYEG/Sec 61 complex and YidC/Oxa1p/Alb3p, are known to exist ubiquitously. In addition to protein integration that depends on these factors (Sec-dependent integration), a Sec-independent pathway is also known1,2,3,4. To clarify these mechanisms in detail, we have tried to reconstitute protein integration in a model organism, E. coli. We have been successful, the integration reactions in vivo being faithfully reflected, and have identified a novel integration factor, glycolipid MPIase, since our structure determination studies revealed that MPIase is a glycolipid devoid of a proteinaceous moiety5,6,7. Thus, MPIase (Fig. 1a) is involved in both Sec-dependent and -independent protein integration into (proteo)liposomes5,6,7,8. Consistently, an anti-MPIase antibody inhibits protein integration into inner membrane vesicles (INV) prepared from E. coli7, indicating that MPIase functions on the cytoplasmic side of inner membranes. Other than the accumulated lines of evidence of the involvement of MPIase in vitro, there is no evidence that MPIase is involved in protein integration in vivo, due to the lack of information on MPIase biosynthesis.
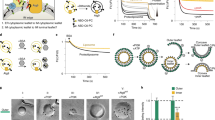
CdsA and YnbB are biosynthetic enzymes for MPIase. (a) Structure of MPIase7. The parts corresponding to compound I (GlcNAc-PP-DAG) and ManNAcA-GlcNAc-PP-DAG are indicated. (b,c) Cytidine deprivation causes MPIase depletion (b) and accumulation of M13 procoat (c). JF618 (pyrG) cells, cultivated in M9 medium supplemented with 250 μg/ml cytidine, were washed three times using fresh M9 medium, followed by inoculation into M9 medium supplemented with 250 μg/ml cytidine (+) or not supplemented (−). At the mid log phase, cells were recovered to analyze the MPIase level by SDS-PAGE/immunoblotting. The SecB level was analyzed as a loading control. To examine the processing of M13 procoat, JF618 harboring pMS119-PC was cultivated as in (b), followed by IPTG (1 mM) induction. Cells were pulse-labeled for 2 min. M13 procoat and coat were immunoprecipitated and visualized by SDS-PAGE/autoradiography. (c,d) Overproduction of CdsA and YnbB causes an increase in the MPIase level. INV were prepared from BL21 (wt), BL21 harboring pT7-CdsA (cdsA++), or BL21 harboring pT7-YnbB (ynbB++). The levels of CdsA (top), MPIase (middle), and SecG (bottom; loading control) were determined by SDS-PAGE/immunoblotting. The relative amounts of the bands are shown. (e) Compound I biosynthesis on the CdsA8 mutant. INV prepared from RS80 (cdsA8 Δcdh) were incubated with [14C]PA, CTP and GlcNAc-phosphate under the specified conditions, followed by TLC analysis. Synthetic references of PA (lane 1), CDP-DAG (lane 2), and compound I (lane 3) were also analyzed. In lane 7, [14C]PA was applied. The references and the sample in lane 6 were co-developed (lanes 9 and 12), while references (lanes 8 and 11) and the sample in lane 6 (lanes 10 and 13) were independently developed, respectively. The spots were visualized with anisaldehyde/H2SO4 (lanes 8–10) or by autoradiography (lanes 11–13). The contaminants in [14C]PA, and are indicated by ‘#’. OG (upper) and sucrose (lower) are by asterisks. (f) LC-MS analysis of in vitro synthesized compound I. (upper) The total ion chromatogram (TIC) from the biosynthetic reaction mixture. (middle) The extracted ion chromatogram (EIC) of m/z 930.5114 from the biosynthetic reaction mixture. (bottom) EIC of m/z 930.5114 from the synthetic reference. (g) The MPIase level in GN80 (cdsA8) increased under non-permissive pH conditions. GN80 grown at pH6.5 was cultivated either at pH6.5 or pH8.5. The levels of MPIase and SecB were determined by SDS-PAGE/immunoblotting.
ECA (enterobacterial common antigen) is a glycolipid found in the outer membranes of Gram-negative bacteria9. Although ECA has the same trisaccharide unit as that of MPIase, knockout of each biosynthetic gene for ECA does not affect MPIase expression7, indicating the presence of an independent set of genes for MPIase biosynthesis. The structural differences between MPIase and ECA are the lengths of the glycan, and the linker between the glycan chain and the lipid moiety7,9. The numbers of repeating trisaccharide units in ECA are more heterogeneous and greater (n = 18~55), while that in MPIase is 9~11. The linker for ECA is monophosphate, while that for MPIase is pyrophosphate. This difference in the linker may reflect the differences in the biosynthetic pathways. Although the glycan unit for ECA is biosynthesized on a carrier lipid, undecaprenol10, depletion of 4-acetamido-4-deoxyfucose (Fuc4NAc) resulted in the accumulation of not only ManNAcA-GlcNAc-PP-undecaprenol but also ManNAcA-GlcNAc-PP-DAG (Fig. 1a); referred to as “DGP-disaccharide’11. This finding strongly suggests that ‘DGP-disaccharide’ is a possible intermediate for MPIase, and that MPIase is biosynthesized on phosphatidic acid, i.e., the first step for MPIase biosynthesis is generation of GlcNAc-PP-DAG (compound I, Fig. 1a). Therefore, we tried to identify the enzyme that catalyzes the generation of compound I, and found that CdsA, a CDP-DAG synthase involved in phospholipid biosynthesis12,13, is also involved in compound I biosynthesis. The combination of CdsA depletion and yeast Tam41p (a mitochondrial CDP-DAG synthase)14 expression revealed that MPIase is essential for membrane protein integration and therefore for cell growth. We also found that Cds1p, a eukaryotic homologue of CdsA15,16, has the ability to biosynthesize MPIase, suggesting that an MPIase homologue is conserved among eukaryotic cells.
Results
CdsA biosynthesizes compound I (GlcNAc-PP-DAG)
We assumed that the first pathway of MPIase biosynthesis is the generation of compound I. In the course of a search for biosynthetic substrates, we found that cytidine deprivation, with a cytidine auxotroph, JF618 (pyrG51)17, caused a reduction in the MPIase level detectable on immunoblotting with anti-MPIase antibody (Fig. 1b), suggesting that cytidinated compounds such as CDP-diacylglycerol (CDP-DAG) are utilized for MPIase biosynthesis. Under the cytidine deprivation conditions, accumulation of M13 procoat (precursor of the major coat protein of M13 bacteriophage) was also observed, strongly suggesting that MPIase is involved in protein integration in vivo (Fig. 1c). Upon membrane integration, M13 procoat undergoes processing of its signal (or leader) sequence, yielding mature M13 coat (Supplementary Fig. 1a). We assumed that compound I is generated through CDP-DAG, a precursor of all the phospholipids in E. coli18, of which biosynthesis is catalyzed by a CDP-DAG synthase, CdsA13,19. Upon CdsA overproduction, we observed a ~3-fold increase in both the CdsA and MPIase levels (Fig. 1d), suggesting that CdsA is involved in MPIase biosynthesis. YnbB is a paralog of CdsA (Supplementary Fig. 2). When YnbB was overexpressed, a similar increase in the MPIase level was also observed (Fig. 1d), indicating that YnbB possesses similar functions to CdsA.
An attempt to biosynthesize compound I in vitro by incubating CTP, PA and some possible GlcNAc donors [UDP-GlcNAc, CDP-GlcNAc and GlcNAc-1-phosphate (GlcNAc-P)] in the presence of INV and/or cytosol prepared from wild type E. coli cells was unsuccessful, presumably reflecting the very low level of MPIase expression5. On the other hand, when INV prepared from CdsA-overproducing cells were incubated with [14C]PA, CTP and a GlcNAc donor, we observed a compound similarly migrated on TLC to the synthetic reference of compound I (a brown spot) in a GlcNAc-P-dependent manner (Supplementary Fig. 3, Rf value = ~0.44 with Solvent system A). The compound was developed at the same position as the synthesized reference of compound I with two different kinds of solvent systems (see below). GN80 (cdsA8) is a pH-sensitive mutant of cdsA that accumulates PA under high pH conditions20. To monitor both compound I and CDP-DAG generation, we constructed RS80, a Cdh (CDP-DAG hydrolase)-deficient version of GN80. When INV prepared from RS80 were incubated with [14C]PA, CTP and GlcNAc-P, compound I was generated (Fig. 1e, lane 6, Rf value = ~0.24 with Solvent system B). The amount of compound I increased while that of CDP-DAG (Rf value = ~0.16) decreased, as the pH of the reaction mixture increased (Fig. 1e, lanes 4~6). The lower and upper products in lanes 4–6 migrated to the positions of synthetic references for CDP-DAG (lane 2) and compound I (lane 3), respectively. The identity of the generated compound I was further confirmed by co-development of the sample in lane 6 and the synthetic references (lanes 9 and 12), since the radioactive compound I and the synthetic reference for compound I showed a perfect match. Time course analysis revealed that compound I biosynthesis occurred mainly at high pH, while CDP-DAG was synthesized at pH 6.5 (Supplementary Fig. 4). The formation of compound I was dependent on CTP, GlcNAc-P, and INV (Supplementary Fig. 5). Immediately after the addition of GlcNAc-P during the biosynthesis reaction, compound I was generated (Supplementary Fig. 6). The structure of the compounds generated in the in vitro reactions involving the CdsA8 mutant were confirmed by LC-mass spectrometry (MS). Figure 1f shows a mass spectrum of the extract from the biosynthesis reaction mixture. The reaction mixture gave an [M-H]− peak originated from compound I (m/z value of 930.5) at the retention time of 5.2 min, which is the same as that of the synthetic reference (Fig. 1f). The peak at 5.2 min included mainly two materials, of which the exact mass numbers were m/z 930.5131 and 952.5047, respectively. The former corresponded to compound I (C43H82NO16P2−, theoretical value 930.5114) and the latter corresponded to CDP-DAG (C44H80N3O15P2−, theoretical value 952.5070) (Supplementary Fig. 7). Moreover, MS/MS analysis from the former peak gave two fragment ions (m/z 709.4155 and 362.0046), which are identical with those from the synthetic reference (Supplementary Fig. 8). The peak of compound I was not observed when GlcNAc-P was omitted in the reaction mixture, while CDP-DAG was generated under the condition. (Supplementary Fig. 9). Partially purified CdsA8 was subjected to the reaction for compound I biosynthesis (Supplementary Fig. 10). Under these high pH conditions, the level of compound I increased with the reaction time, while that of CDP-DAG remained constantly low throughout the reaction time. The level of CDP-DAG (0.7~1.4 pmol/20 μL reaction) was similar to that of CdsA8 (0.8~1.6 pmol/20 μL reaction). These observations strongly suggest that CDP-DAG is synthesized and then converted to compound I on CdsA. They also suggest that CDP-DAG is not released from the CdsA8 mutant, unless it is converted to compound I. Consistent with the efficient generation of compound I at high pH by the CdsA8 mutant, we observed an increase in the MPIase level when GN80 was cultivated at a non-permissive pH (Fig. 1g). Sequencing of the cdsA8 allele revealed that this mutation comprises an amino acid substitution of Tyr 207 to His, suggesting that deprotonation of the His residue at the mutation point impairs the CdsA function in CDP-DAG biosynthesis but not that in compound I biosynthesis.
CdsA depletion causes MPIase depletion with accumulation of M13 procoat
We then constructed knockout mutants of cdsA and ynbB to determine whether or not CdsA and/or YnbB are involved in MPIase biosynthesis. The cdsA gene is the second one in the ispU-cdsA-rseP-bamA operon (Fig. 2a), all of them being essential, while ynbB is not essential21. In order not to disturb the gene expression of the operon, the open reading frame of cdsA was replaced with that of the cat gene (Supplementary Fig. 11) in the presence of the complementary plasmid pAra-CdsA, in which cdsA is placed under the control of the ara regulon (Fig. 2a). The ynbB knockout grew as well as the parent strain (Fig. 2b and Supplementary Fig. 12). On the other hand, both the cdsA knockout and the cdsA/ynbB double knockout were unable to grow in the absence of inducer arabinose (Fig. 2b and Supplementary Fig. 12), confirming that cdsA is essential. We found that MPIase was hardly detected in both the cdsA and cdsA/ynbB knockouts, when CdsA was depleted by cultivating them in the medium without arabinose (Fig. 2c, top). In the ynbB knockout, the MPIase level was not significantly changed (EK1406), indicating that the YnbB effect is marginal or auxiliary. These observations indicate that CdsA is a biosynthetic enzyme for MPIase. Next, we examined membrane integration of M13 procoat under the CdsA- and CdsA/YnbB-depleted conditions. When CdsA or CdsA/YnbB were depleted (i.e., when MPIase was depleted), accumulation of M13 procoat was observed, while M13 procoat was efficiently processed into the mature form in the presence of CdsA (Fig. 2c, bottom). Pulse-chase experiments revealed that upon MPIase depletion, M13 procoat was never processed into the mature form, but was quickly degraded (Fig. 2d,e). These observations strongly suggest that MPIase functions as a membrane protein integrase in vivo.
MPIase is essential for membrane protein integration in vivo. (a) Genetic organization of the cdsA and/or ynbB knockout mutants. In the ΔcdsA strains, CdsA is expressed from ParaB-cdsA on plasmid pAra-CdsA. (b) Growth of the cdsA and/or ynbB knockout mutants. The indicated strains were streaked onto LB plates supplemented with 0.2% arabinose or not supplemented, and then incubated at 37 °C for 16 h. (c) MPIase depletion and M13 procoat accumulation upon CdsA depletion. Overnight cultures of the indicated strains harboring pMS119-PC were washed three times with fresh LB medium, followed by cultivation in the presence and absence of 0.2% arabinose at 37 °C. When the growth of the cdsA knockouts ceased, 1 mM IPTG was added to induce M13 procoat, and cultivation was continued for 1 h. Total cellular proteins were precipitated with TCA, followed by SDS-PAGE/immunoblotting using anti-MPIase (upper) and anti-M13 coat (lower) antisera. (d,e) M13 procoat is accumulated on MPIase depletion. Overnight cultures of KS22 and KS23 harboring pMS119-PC were washed three times with fresh M9 medium, followed by cultivation in the presence (CdsA+/YnbB+ and CdsA+/ΔYnbB) and absence (ΔCdsA/YnbB+ and ΔCdsA/ΔYnbB) of 0.2% arabinose, respectively. After 5-min induction with 1 mM IPTG, they were pulse-labeled for 30 s and then chased for the indicated periods. M13 procoat and coat were immunoprecipitated, followed by SDS-PAGE/autoradiography. The percentage of M13 procoat, obtained in (d), was determined and plotted against chase time (e).
MPIase is essential for cell growth and protein integration
CdsA synthesizes CDP-DAG, a precursor of all phospholipids in E. coli13,20,22, which should be necessary for cell growth. To determine the essentiality of MPIase, we introduced mitochondrial Tam41p of yeast23 into the cdsA/ynbB knockout. Tam41p is a CDP-DAG synthase but exhibits no sequence homology to CdsA/YnbB14. When TAM41 lacking the mitochondrial targeting signal14 was expressed in the cdsA or cdsA/ynbB knockout, PA accumulation caused by CdsA depletion was significantly relieved (Fig. 3a), indicating that Tam41p suppressed the defect in phospholipid biosynthesis. Functional expression of Tam41p in E. coli was also confirmed with the CdsA8 mutant. When CdsA8 was expressed in the cdsA knockout, no growth at high pH was observed (Fig. 3b). Under these conditions, severe accumulation of PA was observed (Fig. 3c). This growth defect and PA accumulation were clearly suppressed by Tam41p expression (Fig. 3b,c).
MPIase is essential for cell growth. (a) Accumulation of PA is relieved on Tam41p induction. The indicated cells harboring plasmids pAra-CdsA and RKP153 (for Tam41p expression from the T7 promoter), cultivated in LB medium in the presence and absence of 0.2% arabinose (for CdsA induction) or 0.1 mM IPTG (for Tam41p induction), were labeled with [14C] palmitic acid. Phospholipids were analyzed by TLC and autoradiography (top). Each reference was analyzed at the right. PE, phosphatidylethanolamine; PG, phosphatidylglycerol; CL, cardiolipin. The PA content in phospholipids was determined (bottom). (b) The pH-sensitive cdsA8 mutation was complemented by Tam41p expression. EK23 (ΔcdsA) cells transformed with either pTet-CdsA (wt) or pTet-CdsA8 (CdsA8) with (right) or without (left) Tam41p expression (by pTac-Tam41p), were streaked onto LB plates at pH 8.5. (c) PA accumulation by cdsA8 was relieved by Tam41p expression. Phospholipids in the indicated cells were analyzed by TLC and visualized with anisaldehyde/H2SO4. (d) Tam41p expression does not restore MPIase expression. The MPIase level in cells cultivated as in (a) was determined by immunoblotting. (e) MPIase is essential for cell growth. KS44 and KS46 harboring plasmids pAra-CdsA and RKP153 were streaked onto LB plates supplemented with 0.2% arabinose (for CdsA induction) or 0.1 mM IPTG (for Tam41p induction), and then incubated at 37 °C for 16 h. (f,g) M13 procoat is accumulated on MPIase depletion even in the presence of Tam41p. KS22 and KS23 cells harboring plasmids pAra-CdsA, pTet-Tam41p and pMS119-PC were subjected to pulse-chase experiments, as described in Fig. 2d. M13 procoat and coat were immunoprecipitated, followed by SDS-PAGE/autoradiography. The percentage of procoat, obtained in (f), was determined (g).
In the ΔcdsA strain, MPIase depletion was rather more complete upon Tam41p expression (Fig. 3d), indicating that Tam41p synthesizes CDP-DAG but not compound I. Under the MPIase-depleted conditions with Tam41p expression, the cdsA and cdsA/ynbB knockouts were unable to grow (Fig. 3e and Supplementary Fig. 13), indicating that MPIase is essential for cell growth.
M13 procoat accumulation was also observed under the MPIase-depleted conditions when Tam41p was expressed (Supplementary Fig. 14). Pulse-chase experiments again demonstrated that M13 procoat was never converted into the mature form but was quickly degraded upon MPIase depletion (Fig. 3f,g), indicating that MPIase is essential for M13 procoat integration in vivo. Unlike the cells without Tam41p expression (Fig. 2e), we observed procoat accumulation in the absence of YnbB (Fig. 3f,g), consistent with the finding that Tam41p overexpression renders MPIase depletion more complete (Fig. 3d).
MPIase stimulates preprotein translocation in vivo
MPIase stimulates preprotein translocation5,24 by affecting the dimer structure of protein translocon SecYEG24. We examined the effect of MPIase depletion on preprotein translocation in vivo (Fig. 4). Processing of the signal (or leader) sequence of a presecretory protein, OmpA, was monitored as an index of preprotein translocation25 (Supplementary Fig. 1b) by means of pulse-chase experiments. We observed accumulation of pOmpA upon MPIase depletion, while most pOmpA materials were chased into mature OmpA in 4 min (Fig. 4a,b). Comparison of the half-life of pOmpA revealed that CdsA/YnbB depletion caused a ~10-fold decrease in the translocation rate (Fig. 4e), consistent with the results obtained in the in vitro reconstitution study24. Induction of Tam41p, which renders MPIase depletion more complete (Fig. 3d), caused more severe accumulation of pOmpA (Fig. 4c–e). Even in the absence of YnbB, pOmpA accumulation was observed, as seen for that of M13 procoat (Fig. 3f,g). These observations indicate that MPIase stimulates preprotein translocation, as revealed in the in vitro analysis5,24.
MPIase stimulates preprotein translocation. (a) pOmpA is accumulated on MPIase depletion. KS22 and KS23 cells were cultivated as described in the legend to Fig. 2d, except that IPTG induction was omitted. Cells were then pulse-labeled for 30 s, and then chased for the indicated periods. OmpA and pOmpA were immunoprecipitated, followed by SDS-PAGE/autoradiography. (b) The results in (a) were quantitated and plotted against chase time. (c) pOmpA is also accumulated on MPIase depletion in the presence of Tam41p. KS22 and KS23 cells harboring pTet-Tam41p were subjected to pulse-chase experiments as in (a). (d) The results in (c) were quantitated and plotted against chase time. (e) Summary of the pulse-chase experiments. The half-lives of pOmpA in (b,d) were calculated.
INV prepared from MPIase-depleted cells are defective in protein integration and preprotein translocation
Next, INV were prepared from MPIase-depleted cells, followed by in vitro integration assaying (Fig. 5). We observed efficient depletion of MPIase (Fig. 5a). Under the conditions for the MPIase depletion, the expression levels of some membrane proteins including YidC26, SecD27 and LolC28 remained unchanged (Fig. 5a). On the other hand, the SecA level increased upon MPIase depletion (Fig. 5a), consistent with the previous reports that SecA is induced when protein translocation is inhibited29,30. We then examined whether or not membrane integration of M13 procoat is inhibited in vitro upon MPIase depletion, as observed in vivo. To avoid complexity due to signal (or leader) processing, we used M13 procoat H5 with an uncleavable signal (or leader) sequence31. We observed a severe defect in M13 procoat H5 integration into MPIase-depleted INV, as reflected by the disappearance of the membrane-protected fragment (MPF)5,32 (Fig. 5b). The integration level for ΔMPIase INV was nearly the same as that in the absence of INV, indicating that M13 procoat integration is dependent on MPIase. These observations are consistent with the in vivo results (Figs 2 and 3). Essentially the same results were obtained when 3L-Pf3 coat (Fig. 5c) and the F0c subunit of F0F1-ATPase (Fig. 5d, Supplementary Fig. 15) were synthesized, confirming the previous findings on reconstitution7,8,33. Thus, that MPIase functions as a membrane protein integrase has been demonstrated both in vitro and in vivo. When pOmpA translocation was examined, we observed significant stimulation of pOmpA translocation in the presence of MPIase (Fig. 5e), again confirming the in vivo results (Fig. 4).
In vitro protein integration activity is impaired upon MPIase depletion. (a) The levels of some membrane proteins do not change upon MPIase depletion. INV were prepared from KS23 harboring plasmid pAra-CdsA cells cultivated in the presence or absence of 0.2% arabinose (cdsA+), followed by SDS-PAGE/immunoblotting using antisera against MPIase or the specified proteins. The relative amount as to the sample ‘+’ is indicated at the bottom of each gel. In the case of SecA, proteins from whole cells were applied. (b–e) Protein integration and translocation into ΔMPIase INV are impaired. Membrane proteins (M13 procoat (b), 3L-Pf3 coat (c), and F0c (d)), and presecretory protein pOmpA (e) were synthesized in vitro by means of the pure system in the presence of +MPIase and ΔMPIase INV. pOmpA gave several bands as structurally different forms53,54. After 30-min incubation at 37 °C, the reaction mixture was divided into two parts; one was analyzed as a translation control of 20% in the ‘−PK’ lanes, and the other was digested with proteinase K (PK) (+). At right, INV were not added (−). The integration and translocation activities are indicated at the bottom of each gel. In (d), a space for the duplicated sample (Supplementary Fig. 13) was removed.
Cds1p, a eukaryotic homolog of CdsA, can biosynthesize MPIase
CdsA is ubiquitously conserved in all organisms. The eukaryotic homologue of CdsA is Cds1p (Supplementary Fig. 16). Next, we examined whether or not eukaryotic Cds1p can substitute for CdsA in the biosynthesis of MPIase. The CDS1 gene of S. cerevisiae (Sc-CDS1) was introduced in the ΔcdsA knockout. Although Sc-Cds1p expression could not complement ΔcdsA, co-expression of Sc-Cds1p and Tam41p allowed the cell growth of the ΔcdsA knockout (Fig. 6a), suggesting that Sc-Cds1p biosynthesizes a sufficient amount of compound I but not of CDP-DAG. Immunoblot analysis confirmed that Sc-Cds1p synthesized MPIase when Tam41p reinforced the CDP-DAG synthase activity (Fig. 6b). Pulse-chase experiments (Fig. 6c,d) revealed that pOmpA processing was recovered to the control level (ΔCdsA/CdsA+) when both Sc-Cds1p and Tam41p were expressed in the ΔcdsA knockout, while only Tam41p expression did not suppress the translocation defect of pOmpA, indicating that Cds1p is responsible for the biosynthetic activity of MPIase. Under these conditions, the MPIase level induced by Sc-Cds1p and that by plasmid-encoded CdsA were similar, but these were lower than that in wt cells (Fig. 6e), explaining the partial recovery of pOmpA translocation (Fig. 6c). Essentially the same results were obtained using human Cds1p (Hs-Cds1p), i.e., Hs-Cds1p relieved the growth of the ΔcdsA knockout (Fig. 6a) and MPIase expression (Fig. 6b,e) together with Tam41p. Protein translocation (Fig. 6c,d) was also recovered on Hs-Cds1p expression together with Tam41p. Thus, a eukaryotic homolog Cds1p could be involved in MPIase biosynthesis.
Eukaryotic Cds1p can synthesize MPIase. (a) Both Sc-Cds1p and Hs-Cds1p complement ΔcdsA when combined with Tam41p. KS22 cells harboring plasmids pAra-CdsA and pTac-Tam41p transformed with either pACYC-Km (top), pTet-Cds1p-Sc (middle), or pTet-Cds1p-Hs (bottom) were streaked onto LB plates supplemented with the specified inducer, followed by incubation at 37 °C for 16 h. (b) Both Sc-Cds1p and Hs-Cds1p restored the MPIase level when combined with Tam41p. The MPIase level in KS22 cells, in which the specified proteins had been induced, was determined by immunoblotting (top). The SecB level was also determined as a loading control (bottom). (c,d) pOmpA translocation was relieved on both Cds1p and Tam41p expression in the absence of CdsA. KS22 cells harboring plasmids pAra-CdsA and pTac-Tam41p plus either pACYC-Km, pTet-Cds1p-Sc, or pTet-Cds1p-Hs were pulse-labeled for 30 s and then chased for the indicated periods, as described in the legend to Fig. 4. pOmpA and OmpA were immunoprecipitated, followed by SDS-PAGE and autoradiography. EK413 (wt) cells were also analyzed as a CdsA+ control. The results in (c) were quantitated and plotted against chase time (d).
Discussion
In this paper, we have demonstrated that MPIase is essential for membrane protein integration in vivo and therefore for cell growth. Moreover, we have shown that MPIase stimulates preprotein translocation in vivo. This was achieved through the finding that CdsA is involved in MPIase biosynthesis, in addition to the well-defined function of a CDP-DAG synthase20,22, by which all the phospholipids in E. coli are biosynthesized18. The possibility that a defect in phospholipid biosynthesis caused by CdsA depletion perturbs membranes, which causes pleiotropic effects including the impairment of protein integration, could be excluded due to the result of introduction of Tam41p, which synthesizes CDP-DAG but not compound I. Moreover, the membrane integrity was maintained under our experimental conditions, irrespective of Tam41p induction, since pOmpA was finally processed into mature OmpA even under the MPIase depletion conditions (Fig. 4). Although MPIase-depleted cells should be lysed after prolonged cultivation, the levels of some membrane proteins remained unchanged under our experimental conditions (Fig. 5a). Although we could not rule out a malfunction of other integration factors that might indirectly cause a defect in protein integration, such effects would nevertheless be caused by MPIase depletion. Thus, all the findings that we have made by means of the in vitro reconstitution system as to MPIase5,6,7,8,24 were supported in vivo.
We assumed that the initial step of MPIase biosynthesis is the formation of compound I, since a possible biosynthetic intermediate for MPIase, ManNAcA-GlcNAc-PP-DAG (‘DGP-disaccharide’), has been identified upon depletion of Fuc4NAc11. This compound was identified during the course of identification of a biosynthetic intermediate of ECA, of which the glycan is similar to that of MPIase7, but it turned out not to be an intermediate for ECA11. For ECA biosynthesis, the repeating unit of the glycan composed of three N-acetylated sugars is synthesized on a carrier lipid, undecaprenol, followed by polymerization of the glycan chain9. On the other hand, the identification of ManNAcA-GlcNAc-PP-DAG (‘DGP-disaccharide’) strongly suggests that the repeating unit for MPIase is synthesized on PA to allow its subcellular localization at the inside of inner membranes7, while the mechanism underlying glycan polymerization is totally unknown. Since the knockout mutant as to each gene for ECA biosynthesis still expressed MPIase at the wild-type level7, E. coli should possess a complete set of biosynthetic genes for MPIase. The cdsA gene is one of such MPIase-specific ones. We demonstrated that functionally expressed Tam41p suppressed the defect in phospholipid biosynthesis but not the defect in MPIase biosynthesis. These observations indicate that biosynthesis of CDP-DAG is not sufficient for compound I biosynthesis. On the other hand, the decrease in MPIase level in the pyrG mutant revealed that cytidine nucleotides are required for MPIase biosynthesis. Compound I biosynthesis, which was confirmed by MS analysis (Fig. 1f and Supplementary Figs 7 and 8), was dependent not only on GlcNAc-P but also on CTP in the in vitro reactions. Moreover, in the reactions involving an extract containing the pH-sensitive mutant CdsA820, CDP-DAG was initially biosynthesized to the level of CdsA8, and subsequently compound I was generated, suggesting that CDP-DAG was converted into compound I through the incorporation of GlcNAc-P. The observation that nucleotide sugars such as UDP-GlcNAc and CDP-GlcNAc were not incorporated into PA or CDP-DAG yielding compound I (Supplementary Fig. 3) is consistent with the assumption that GlcNAc-P substitutes for the CMP moiety of CDP-DAG. MS analysis also demonstrated the generation of compound I in a GlcNAc-P-dependent manner (Supplementary Fig. 9). Considering that Tam41p could not biosynthesize compound I, this substitution should occur on CdsA (Fig. 7). The cdsA8 mutant required co-expression of Tam41p for growth because CDP-DAG would hardly dissociate from CdsA, and a sufficient amount of CDP-DAG for the biosynthesis of bulk phospholipids was not provided. On the other hand, under the non-permissive conditions, MPIase was expressed at a higher level than in the wild-type, suggesting that low dissociation of CDP-DAG would be advantageous for GlcNAc-P incorporation (Fig. 7). The possibility cannot be excluded that some unknown factor(s) are necessary for GlcNAc-P incorporation. Nonetheless, the expression level of MPIase is quite low compared with that of bulk phospholipids; the MPIase level could be estimated to be <1/1,000 of that of phospholipids as a molar ratio5,6,7. MPIase has a labile pyrophosphate as a linkage between glycans and lipids7, while ECA has a monophosphate linkage9. This difference suggests that the presence of pyrophosphate could be involved in the control of MPIase level to keep it low, unlike ECA. The low level of MPIase expression is the possible reasons why we could not detect compound I formation under the wild-type conditions. In the cdsA8 background, retardation of the release of CDP-DAG greatly helped to detect compound I. According to the crystal structure of CdsA34, the mutation point in the cdsA8 allele (Tyr207His) lies in transmembrane stretch 6, which is close to the substrate-binding site. Under the non-permissive conditions at high pH, the His residue at the mutation point should be deprotonated, and therefore the mutation would stabilize the interaction between CdsA and CDP-DAG, thereby retarding the release of CDP-DAG (Fig. 7).
Working model for the CdsA/YnbB function with respect to MPIase biosynthesis. Both CdsA/YnbB and Tam41p biosynthesize CDP-DAG from CTP and PA. Unlike Tam41p, CdsA/YnbB also biosynthesize compound I immediately after CDP-DAG biosynthesis. The cdsA8 mutation may retard the release of CDP-DAG from CdsA, which results in an increase in the efficiency of compound I biosynthesis.
YnbB is a paralog of CdsA with a homologous region in the C-terminal half (Supplementary Fig. 2), while both YnbB and CdsA possess 8~9 transmembrane stretches. YnbB homologs are widely conserved among bacteria, although YnbB is not essential for growth. Since YnbB could not complement the ΔcdsA knockout even upon YnbB overproduction (data not shown), but its overproduction resulted in MPIase overproduction as well as CdsA overproduction, the YnbB function might be a backup for MPIase biosynthesis, but not for phospholipid biosynthesis. In this regard, the N-terminal half of CdsA might accelerate CDP-DAG biosynthesis of bulk phospholipids, while the role of the N-terminal half of YnbB is totally unknown. Similarly, Cds1p could biosynthesize a sufficient amount of MPIase but not of bulk phospholipids. Therefore, both YnbB and Cds1p have the ability to biosynthesize CDP-DAG and then compound I, however, the level of CDP-DAG biosynthesis is not sufficient for that of bulk phospholipids. In the presence of Tam41p expression, the effect of ΔynbB was exaggerated; i.e., precursors of M13 coat and OmpA accumulated in the ΔynbB and Tam41p+ background. These observations indicate that CdsA and YnbB compete with Tam41p for PA, resulting in a decrease in the MPIase level (Fig. 7). This competition is consistent with the finding that Tam41p overexpression renders MPIase depletion more complete (Fig. 3d).
MPIase co-precipitates with SecYEG, strongly suggesting their direct interaction24. Since YidC, which also interacts with SecYEG35, is involved in Sec-independent integration including M13 procoat integration26,36, it is likely that MPIase and YidC function in a concerted manner, suggesting that these factors directly interact. Recently, such collaboration between MPIase and YidC was clarified in the case of integration of F0c, the c subunit of F0F1 ATPase8. The MPIase function was clearly demonstrated in the in vitro analysis, which reflects an early stage of protein integration, since the appearance of MPF is used as an index of integration5. On the other hand, it is difficult to analyze the role of YidC in the in vitro system5,33, although YidC depletion causes a general defect in protein integration26. Based on the crystal structure of YidC, a detailed mechanism underlying YidC-dependent integration is proposed, where a hydrophilic cavity inside the membrane is important for integration of membrane proteins37,38. Taking these and the findings in the present study together, we propose that MPIase first drives an initial stage of integration, followed by transfer of membrane proteins to YidC to complete the integration reaction at a late stage. This model is consistent with the observation that upon YidC depletion M13 procoat is accumulated as an alkaline extraction-resistant form36. Under the conditions, M13 procoat integration proceeds with the MPIase function, but is not completed because of the lack of the YidC function, the precursor form remaining unprocessed.
Homologues of CdsA are ubiquitously conserved as Cds1p in eukaryotic cells15,39. If GlcNAc-P is sufficient for compound I synthesis without the requirement of additional factors, eukaryotic cells can synthesize compound I, since GlcNAc-P is also ubiquitously present. Although sugars in MPIase other than GlcNAc (Fuc4NAc and ManNAcA) seem specific in bacteria, and compound I might be an intermediate of a glycolipid with unknown functions, it is plausible that a compound homologous to MPIase, which is involved in membrane protein integration, is also ubiquitously conserved. Identification of such a homologue would shed new light on the molecular mechanisms underlying membrane protein integration in eukaryotic cells including those of higher plants and animals.
Methods
Materials
The E. coli strains (Table S1), plasmids (Table S2), and PCR primers (Table S3) used in this study are listed. SecA40 and SecB41 were purified as described. Antibodies against MPIase7, SecB42, SecD27, SecG43 and OmpA44 were raised in rabbits using the respective purified factors, while those against CdsA (Glu75~Ser92), M13 procoat (Ala24~Tyr44), LolC (Lys239~Glu255)28, and YidC (Lys382~Gln402) were raised against synthetic peptides corresponding to the specified regions. All the antibodies were obtained through the commercially available custom services. [Glycerol-14C(U)]-L-α-dipalmitoyl-phosphatidic acid (3.7~7.4 GBq/mmol), [14C(U)] palmitic acid (>18.5 GBq/mmol), and [35S] EXPRESS Protein Labeling Mix, a mixture containing both [35S] Met and [35S] Cys (~37 TBq/mmol), were obtained from Perkin Elmer, Inc. CDP-DAG, PA, PG and PE were from Avanti Polar Lipids, Inc. CL, GlcNAc-P, UDP-GlcNAc and L-arabinose were obtained from Sigma-Aldrich. CTP was from Roche Diagnostics. IPTG was from Wako Pure Chemical Industries, Ltd. CDP-GlcNAc (Supplementary Fig. 18) and the authentic reference of compound I (Supplementary Fig. 19) were chemically synthesized as described under Supplementary Methods. Protein A Sepharose was from GE Healthcare Life Sciences. TLC plates were from Merck.
Biosynthetic formation of compound I
INV (2 mg/mL), prepared from RS80 (cdsA8 Δcdh), were solubilized with 1.5% octyl glucoside, and then mixed with [14C] PA (4 μM; ~23 kBq/mL), 1 mM CTP, 1 mM GlcNAc-P, 5 mM MgSO4 in 30 mM Bistris-H2SO4 (pH 6.5) or 30 mM Hepes-KOH (pH 7.5 or pH8.5), and 1.5% octyl glucoside. In some experiments, a cholate (6%) extract of INV, obtained by centrifugation (170,000 × g, 1 h, 4 °C), was used instead of INV. The reaction mixture (20 μL) was incubated at 37 °C. The reaction was terminated by adding 20 mM EDTA. An aliquot (2.5 μL) was analyzed by TLC using Solvent system B (chloroform/methanol/water/acetic acid: 5/5/0.89/0.11). The radioactive spots were visualized with a Phosphorimager (GE Healthcare Life Science), while a synthetic reference of compound I was stained with anisaldehyde-H2SO4 as described5.
LC-MS analysis
INV (1 mg protein), prepared from RS80/pTac-CdsA8, were solubilized in 1.5% octyl glucoside, 10% glycerol, 1 mM dithiothreitol, 50 mM Tricine-KOH (pH 8.5). Solubilized membrane (0.5 mL), recovered by centrifugation (150,000 × g, 30 min, 4 °C), were mixed with dipalmitoyl-phosphatidic acid at 0.4 mg/mL, followed by dialysis against buffer C (50 mM Tricine-KOH (pH 8.5), 1 mM dithiothreitol) to form proteoliposomes with PA. Proteoliposomes were then recovered by centrifugation (150,000 × g, 60 min, 4 °C), suspended in buffer C at 5 mg protein/mL, and frozen-thawed-sonicated. Proteoliposomes (1.5 mg/mL) thus reconstituted were subjected to the reaction for compound I biosynthesis by incubating with 1 mM CTP, 1 mM GlcNAc-P, 1 mM ATP, 10 mM creatine phosphate, 10 μg/ml creatine kinase, 5 mM MgSO2, 30 mM K2SO4, 20 mM Tricine-KOH (pH 8.5) at 37 °C for 21 h. As a negative control, GlcNAc-P was omitted. After the reaction, proteoliposomes were recovered by centrifugation (150,000 × g, 60 min, 4 °C), and then suspended in MeOH (400 μL). The sonicated suspension was centrifuged (15,000 × g, 25 °C) for 1 min and supernatant (5 μL in each analysis) was subjected to the LC-MS analysis. High-resolution mass spectra of all products including compound I were obtained using an ion-trap time-of-flight mass spectrometer (Shimadzu LCMS-IT-TOF). A COSMOSIL 5C18-AR-II (2.0ID × 150 mm) column was used for separation of enzymatic products. The mobile phase 10 mM AcONH4 aq. (A) and acetone (B) were used for linear gradient elution (0-4-7-7.01-10 min, 60-100-100-60-60% (B)). Positive and negative ions were measured simultaneously. The parameters were as follows: nebulizer gas, 1.5 L/min; drying gas pressure, 190 kPa; CDL temperature, 200 °C; detector, 1.64 kV; and interface, 4.5 kV (positive mode) and −3.5 kV (negative mode).
Construction of the ΔynbB and ΔcdsA strains
Keio clone JW140621 was used as a donor of the ΔynbB::kan allele, which was transduced into EK41343 and BL21 (DE3)45 by means of the P1 phage, yielding EK4106 and BL101, respectively. The kan cassette in EK4106 and BL101 was removed using plasmid pCP2046, as described47, yielding KS21 (EK413 ΔynbB) and KS42 (BL21 (DE3) ΔynbB). The strategy for construction of the ΔcdsA knockout is illustrated in Supplementary Fig. 11. The ΔcdsA::cat allele was introduced by P1 transduction into EK413, KS21 (EK413 ΔynbB), BL21 (DE3)45, and KS42 (BL21 (DE3) ΔynbB), yielding KS22, KS23, KS44, and KS46, respectively. These strains harbored plasmid pAra-CdsA, and were constructed in the presence of 0.2% arabinose.
Pulse-chase experiments
An overnight culture was washed with M9 medium three times to remove arabinose, followed by 1:1000 inoculation into the M9 medium supplemented with 18 amino acids (10 μM each) other than Met and Cys, and 0.01% yeast extract. Where specified, 0.2% arabinose was added to induce CdsA. When cell growth ceased upon CdsA depletion, [35S]-EXPRESS Protein Labeling Mix was added at ~100 kBq/ml to allow labeling for 30 s. Labeling was terminated by adding non-radioactive Met and Cys (12 mM each). At the specified chase time, 0.5 mL of the culture was withdrawn, and cellular proteins were precipitated with 5% TCA. The TCA precipitates were washed with acetone and ether successively, and then solubilized in 50 μL of 50 mM Tris-HCl (pH 7.5), 1% SDS, 1 mM EDTA. The samples were boiled for 3 min, and then diluted with 1 mL of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF). The insoluble materials were removed by centrifugation (10,000 × g, 5 min at 4 °C). Antiserum (2 μL) against either M13 coat or OmpA was added to the supernatant, and the mixture was incubated overnight at 4 °C. The mixture received 20 μL of protein A Sepharose (50% slurry), and then was kept at 4 °C for 60 min. After brief centrifugation, the resin was washed with 0.5 mL of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, and 0.5 mL of 50 mM Tris-HCl (pH 7.5), 1% Triton X-100, successively. The washed resin was suspended in 20 μL of the sample buffer used for SDS-PAGE and then boiled for 3 min. The immunoprecipitated materials were analyzed by SDS-PAGE and visualized with a Phosphorimager.
Phospholipid analysis
An overnight culture, washed three times with LB medium, was inoculated into LB at the dilution of 1:500. When necessary, arabinose (0.2%) was added to induce CdsA. Tam41p was then induced with 0.1 mM IPTG for 30 min when cell growth ceased. Cells were labeled with [14C] palmitic acid (~50 kBq/mL) for 3 h at 37 °C. An aliquot (0.5 mL) of the culture was centrifuged to recover the cells. Phospholipids were then extracted, as described48. Phospholipids were analyzed by TLC using a solvent system (chloroform/methanol/water: 9/5/1) for development, followed by visualization using a Phosphorimager (GE Healthcare Life Science). Non-radioactive samples were visualized with anisaldehyde-H2SO4, as described5.
Assaying of protein integration and translocation in vitro
Substrate membrane protein or presecretory protein was in vitro synthesized by means of a pure system, a reconstituted translation system composed of purified components49. The reaction mixture for the pure system (20 μL) containing INV (0.2 mg protein/mL), plasmids encoding the substrate protein under the T7 promoter, and [35S] methionine and cysteine (~10 MBq/mL) was incubated for 30 min at 37 °C. Cold methionine was omitted from the reaction. After the reaction had been terminated by chilling on ice, the mixture was divided into two parts. One part (3 μL) was treated with 5% trichloroacetic acid to precipitate proteins. The other part (15 μL) was mixed with an equal volume of proteinase K (1 mg/mL), followed by incubation for 20 min at 25 °C. After trichloroacetic acid (5%) had been mixed in, samples were incubated for 5 min at 56 °C to inactivate proteinase K, followed by recovery of proteins by centrifugation (10,000 × g, 5 min at 4 °C). Radioactive materials were separated by SDS-PAGE and then detected by autoradiography. Radioactivity was quantitated by means of ImageQuant software (GE).
Construction of plasmids carrying genes for Cds1p
The CDS1 gene (1.3 kbp) of S. cerevisiae (Sc-CDS1) with a BamHI site at the 5′ end and a KpnI site at the 3′ end was PCR-amplified using a pair of primers (Sc-CDS1-5′ and Sc-CDS1-3′ comp). The gene fragment encoding Sc-CDS1 was digested with BamHI, and then ligated to plasmid pUSI2 cut with BamHI and SmaI, yielding pTac-Cds1p-Sc. The gene fragment encoding Sc-Cds1p, obtained by digesting pTac-Cds1p-Sc with BamHI and SalI, was cloned into the same sites of pACYC-Km, yielding pTet-Cds1p-Sc. The human CDS1 gene (Hs-CDS1) was chemically synthesized, in which rare codons were replaced with major codons in E. coli (Supplementary Fig. 17). Plasmid pTet-Cds1p-Hs, which encodes Hs-CDS1 under the control of the tet promoter, was constructed as described in the Supplementary Information.
Other methods
SDS-PAGE was carried out using 12.5% acrylamide-0.27% N,N′-bismethyleneacrylamide containing 6 M urea for M13 (pro)coat, 3L-Pf3 coat and F0c, or not containing it for SecB, SecG and MPIase, as described50. Gels composed of 12.5% acrylamide-0.33% N,N′-bismethyleneacrylamide were also used51 to analyze other proteins. Bands on immunoblots were quantitated by means of a CS analyzer (ATTO). Proteins were quantitated using bovine serum albumin as a standard, as described52.
References
Dalbey, R. E., Wang, P. & Kuhn, A. Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 (2011).
Denks, K. et al. The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol. Membr. Biol. 31, 58–84 (2014).
Nishiyama, K. & Shimamoto, K. Glycolipozyme membrane protein integrase (MPIase): recent data. Biomol. Concepts 5, 429–438 (2014).
Endo, Y. & Nishiyama, K. Relationship between glycolipozyme MPIase and components comprising the protein transport machinery. Med. Res. Archives 2, 11 (2015).
Nishiyama, K. et al. A novel complete reconstitution system for membrane integration of the simplest membrane protein. Biochem. Biophys. Res. Commun. 394, 733–736 (2010).
Nishiyama, K. et al. A derivative of lipid A is involved in signal recognition particle/SecYEG-dependent and -independent membrane integrations. J. Biol. Chem. 281, 35667–35676 (2006).
Nishiyama, K. et al. MPIase is a glycolipozyme essential for membrane protein integration. Nat. Commun. 3, 1260 (2012).
Nishikawa, H., Sasaki, M. & Nishiyama, K. Membrane insertion of F0 c subunit of F0F1 ATPase depends on glycolipozyme MPIase and is stimulated by YidC. Biochem. Biophys. Res. Commun. 487, 477–482 (2017).
Rick, P. D. & Silver, R. P. Enterobacterial Common Antigen and Capsular Polysaccharides in Escherichia coli and Salmonella, Cellular and Molecular Biology, 2nd edition, Vol. 1. (ed. Neidhardt, F. C.) 104–122 (ASM Press, Washington, DC 1996).
Rush, J. S., Rick, P. D. & Waechter, C. J. Polyisoprenyl phosphate specificity of UDP-GlcNAc:undecaprenyl phosphate N-acetylglucosaminyl 1-P transferase from E. coli. Glycobiology 7, 315–322 (1997).
Rick, P. D. et al. Characterization of the lipid-carrier involved in the synthesis of enterobacterial common antigen (ECA) and identification of a novel phosphoglyceride in a mutant of Salmonella typhimurium defective in ECA synthesis. Glycobiology 8, 557–567 (1998).
Icho, T., Sparrow, C. P. & Raetz, C. R. Molecular cloning and sequencing of the gene for CDP-diglyceride synthetase of Escherichia coli. J. Biol. Chem. 260, 12078–12083 (1985).
Sparrow, C. P. & Raetz, C. R. Purification and properties of the membrane-bound CDP-diglyceride synthetase from Escherichia coli. J. Biol. Chem. 260, 12084–12091 (1985).
Tamura, Y. et al. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 17, 709–718 (2013).
Shen, H., Heacock, P. N., Clancey, C. J. & Dowhan, W. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J. Biol. Chem. 271, 789–795 (1996).
Weeks, R. et al. Isolation and expression of an isoform of human CDP-diacylglycerol synthase cDNA. DNA Cell Biol. 16, 281–289 (1997).
Friesen, J. D. et al. Isolation of a lambda transducing bacteriophage carrying the relA gene of Escherichia coli. J. Bacteriol. 127, 917–922 (1976).
Raetz, C. R. Molecular genetics of membrane phospholipid synthesis. Annu. Rev. Genet. 20, 253–295 (1986).
Icho, T., Bulawa, C. E. & Raetz, C. R. Molecular cloning and sequencing of the gene for CDP-diglyceride hydrolase of Escherichia coli. J. Biol. Chem. 260, 12092–12098 (1985).
Ganong, B. R. & Raetz, C. R. Massive accumulation of phosphatidic acid in conditionally lethal CDP-diglyceride synthetase mutants and cytidine auxotrophs of Escherichia coli. J. Biol. Chem. 257, 389–394 (1982).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006 0008 (2006).
Ganong, B. R., Leonard, J. M. & Raetz, C. R. Phosphatidic acid accumulation in the membranes of Escherichia coli mutants defective in CDP-diglyceride synthetase. J. Biol. Chem. 255, 1623–1629 (1980).
Tamura, Y. et al. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 174, 631–637 (2006).
Moser, M., Nagamori, S., Huber, M., Tokuda, H. & Nishiyama, K. Glycolipozyme MPIase is essential for topology inversion of SecG during preprotein translocation. Proc. Natl. Acad. Sci. USA 110, 9734–9739 (2013).
Beckwith, J. & Silhavy, T. J. Genetic analysis of protein export in Escherichia coli. Methods Enzymol. 97, 3–11 (1983).
Samuelson, J. C. et al. YidC mediates membrane protein insertion in bacteria. Nature 406, 637–641 (2000).
Matsuyama, S., Fujita, Y. & Mizushima, S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 12, 265–270 (1993).
Narita, S., Kanamaru, K., Matsuyama, S. & Tokuda, H. A mutation in the membrane subunit of an ABC transporter LolCDE complex causing outer membrane localization of lipoproteins against their inner membrane-specific signals. Mol. Microbiol. 49, 167–177 (2003).
Schatz, P. J. & Beckwith, J. Genetic analysis of protein export in Escherichia coli. Annu. Rev. Genet. 24, 215–248 (1990).
Bieker, K. L., Phillips, G. J. & Silhavy, T. J. The sec and prl genes of Escherichia coli. J. Bioenerg. Biomembr. 22, 291–310 (1990).
Kuhn, A. & Wickner, W. Conserved residues of the leader peptide are essential for cleavage by leader peptidase. J. Biol. Chem. 260, 15914–15918 (1985).
Koch, H. G. et al. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10, 2163–2173 (1999).
Kawashima, Y., Miyazaki, E., Muller, M., Tokuda, H. & Nishiyama, K. Diacylglycerol specifically blocks spontaneous integration of membrane proteins and allows detection of a factor-assisted integration. J. Biol. Chem. 283, 24489–24496 (2008).
Liu, X., Yin, Y., Wu, J. & Liu, Z. Structure and mechanism of an intramembrane liponucleotide synthetase central for phospholipid biosynthesis. Nat. Commun. 5, 4244 (2014).
Scotti, P. A. et al. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 (2000).
Samuelson, J. C. et al. Function of YidC for the insertion of M13 procoat protein in Escherichia coli: translocation of mutants that show differences in their membrane potential dependence and Sec requirement. J. Biol. Chem. 276, 34847–34852 (2001).
Kumazaki, K. et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509, 516–520 (2014).
Kumazaki, K. et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 4, 7299 (2014).
Inglis-Broadgate, S. L. et al. Isolation and characterization of murine Cds (CDP-diacylglycerol synthase) 1 and 2. Gene 356, 19–31 (2005).
Akita, M., Sasaki, S., Matsuyama, S. & Mizushima, S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J. Biol. Chem. 265, 8164–8169 (1990).
Nishiyama, K. & Tokuda, H. Development of a functional in vitro integration system for an integral membrane protein, SecG. Biochem. Biophys. Res. Commun. 390, 920–924 (2009).
Shimizu, H., Nishiyama, K. & Tokuda, H. Expression of gpsA encoding biosynthetic sn-glycerol 3-phosphate dehydrogenase suppresses both the LB- phenotype of a secB null mutant and the cold-sensitive phenotype of a secG null mutant. Mol. Microbiol. 26, 1013–1021 (1997).
Nishiyama, K., Suzuki, T. & Tokuda, H. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell 85, 71–81 (1996).
Tani, K., Tokuda, H. & Mizushima, S. Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli. Effect of membrane energization. J. Biol. Chem. 265, 17341–17347 (1990).
Studier, F. W. & Moffatt, B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 (1986).
Cherepanov, P. P. & Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 (1995).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Shimizu, Y. et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001).
Hussain, M., Ichihara, S. & Mizushima, S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J. Biol. Chem. 255, 3707–3712 (1980).
Laemmli, U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970).
Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 (1951).
Pogliano, J. A. & Beckwith, J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 13, 554–561 (1994).
Nishiyama, K. & Tokuda, H. Preparation of a highly translocation-competent proOmpA/SecB complex. Protein Sci. 19, 2402–2408 (2010).
Acknowledgements
We thank Prof Toshihiko Oka (Rikkyo University) for the discussion, Prof Andreas Kuhn (University of Hohenheim) for the plasmids, Ms Mari Saikudo (Iwate University) for the technical help, and NBRP (NIG, Japan): E.coli and E. coli Genetic Resources at Yale (CGSC) for E. coli strains. The experiments involving radioisotopes were carried out at the Radioisotope Laboratory of Iwate University. This work was supported by JSPS KAKENHI (Grant Numbers 17K13262 to K.F.; 16H06156, 16H00797, 26119704 and 26106003 to Y.Kuruma; 15H05595 to Y.T., 15H05705 and 22227003 to T.E.; 16H02089 to T.U.; 16H01166 to K.Shimamoto; 25291009, 26102703, 26119701, 15KT0073, 16H01374, 16K15083 and 17H02209 to K.N.) and JST CREST (Grant Number JPMJCR12M1 to T.E.).
Author information
Authors and Affiliations
Contributions
K.Sawasato performed the in vivo characterization of MPIase and wrote the manuscript. R.S. and Y.Kamemoto performed the identification of the biosynthetic enzyme for MPIase. H.N. performed the functional analysis of MPIase in vitro. N.I. performed the in vivo analysis involving Cds1p. K.F., T. Y. and K. Shimamoto performed the chemical synthesis and MS analyses, and wrote the manuscript. Y.Kuruma and T.U. supplied the pure system optimized for the protein integration assay. Y.T. and T.E. supervised the experiments involving Tam41p. K.N. designed the experiments, analyzed data, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sawasato, K., Sato, R., Nishikawa, H. et al. CdsA is involved in biosynthesis of glycolipid MPIase essential for membrane protein integration in vivo. Sci Rep 9, 1372 (2019). https://doi.org/10.1038/s41598-018-37809-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-37809-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.