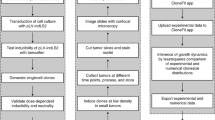
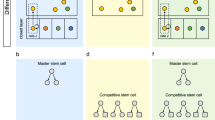
Abstract
The clonogenic assay measures the capacity of single cells to form colonies in vitro. It is widely used to identify and quantify self-renewing mammalian cells derived from in vitro cultures as well as from ex vivo tissue preparations of different origins. Varying research questions and the heterogeneous growth requirements of individual cell model systems led to the development of several assay principles and formats that differ with regard to their conceptual setup, 2D or 3D culture conditions, optional cytotoxic treatments and subsequent mathematical analysis. The protocol presented here is based on the initial clonogenic assay protocol as developed by Puck and Marcus more than 60 years ago. It updates and extends the 2006 Nature Protocols article by Franken et al. It discusses different strategies and principles to analyze clonogenic growth in vitro and presents the clonogenic assay in a modular protocol framework enabling a diversity of formats and measures to optimize determination of clonogenic growth parameters. We put particular focus on the phenomenon of cellular cooperation and consideration of how this can affect the mathematical analysis of survival data. This protocol is applicable to any mammalian cell model system from which single-cell suspensions can be prepared and which contains at least a small fraction of cells with self-renewing capacity in vitro. Depending on the cell system used, the entire procedure takes ~2–10 weeks, with a total hands-on time of <20 h per biological replicate.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Data availability
Source data are provided with this paper. All colony-counting raw data of clonogenic survival experiments in this article (i.e., S-C value pairs of all biological replicates) are provided in the Source Data for Figs. 5 and 9. Some of the clonogenic survival data displayed in Figs. 5 and 9 were taken from ref. 17, as noted in the corresponding figure legends. Schematic graphs in Figs. 2, 4 and 9d were generated from hypothetical datasets. All other data supporting the findings of this study are available within the article and its supplementary information files. Additional information can be provided by the corresponding author upon request.
Code availability
The paper is accompanied by two MS Excel template files for the presented analysis workflows: Supplementary Table 1 for the power regression–based analysis approach and Supplementary Table 2 for PE-based normalization. The R-package CFAcoop is available at https://cran.r-project.org/web/packages/CFAcoop.
References
Franken, N. A., Rodermond, H. M., Stap, J., Haveman, J. & van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319 (2006).
Puck, T. T. & Marcus, P. I. Action of x-rays on mammalian cells. J. Exp. Med. 103, 653–666 (1956).
Puck, T. T., Marcus, P. I. & Cieciura, S. J. Clonal growth of mammalian cells in vitro; growth characteristics of colonies from single HeLa cells with and without a feeder layer. J. Exp. Med. 103, 273–283 (1956).
Puck, T. T. & Marcus, P. I. A rapid method for viable cell titration and clone production with hela cells in tissue culture: the use of X-irradiated cells to supply conditioning factors. Proc. Natl Acad. Sci. USA 41, 432–437 (1955).
Rheinwald, J. G. & Green, H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell 6, 331–343 (1975).
Reynolds, B. A. & Weiss, S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707–1710 (1992).
Phillips, T. M., McBride, W. H. & Pajonk, F. The response of CD24-/low/CD44+ breast cancer-initiating cells to radiation. J. Natl Cancer Inst. 98, 1777–1785 (2006).
Wink, D. A. et al. Nitric oxide protects against cellular damage and cytotoxicity from reactive oxygen species. Proc. Natl Acad. Sci. U. S. A. 90, 9813–9817 (1993).
Liebmann, J. E. et al. Cytotoxic studies of paclitaxel (Taxol) in human tumour cell lines. Br. J. Cancer 68, 1104–1109 (1993).
Wang, Q. et al. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J. Natl Cancer Inst. 88, 956–965 (1996).
Elkind, M. M. & Sutton, H. Radiation response of mammalian cells grown in culture. 1. Repair of X-ray damage in surviving Chinese hamster cells. Radiat. Res. 13, 556–593 (1960).
Huang, P. et al. Folic acid-conjugated silica-modified gold nanorods for X-ray/CT imaging-guided dual-mode radiation and photo-thermal therapy. Biomaterials 32, 9796–9809 (2011).
Elgendy, M., Sheridan, C., Brumatti, G. & Martin, S. J. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol. Cell 42, 23–35 (2011).
Alimova, I. N. et al. Metformin inhibits breast cancer cell growth, colony formation and induces cell cycle arrest in vitro. Cell Cycle 8, 909–915 (2009).
Rochat, A., Kobayashi, K. & Barrandon, Y. Location of stem cells of human hair follicles by clonal analysis. Cell 76, 1063–1073 (1994).
Sanford, K. K., Earle, W. R. & Likely, G. D. The growth in vitro of single isolated tissue cells. J. Natl Cancer Inst. 9, 229–246 (1948).
Brix, N. et al. The clonogenic assay: robustness of plating efficiency-based analysis is strongly compromised by cellular cooperation. Radiat. Oncol. 15, 248 (2020).
Orth, M., Unger, K., Schoetz, U., Belka, C. & Lauber, K. Taxane-mediated radiosensitization derives from chromosomal missegregation on tripolar mitotic spindles orchestrated by AURKA and TPX2. Oncogene 37, 52–62 (2018).
Hess, J. et al. Genomic amplification of Fanconi anemia complementation group A (FancA) in head and neck squamous cell carcinoma (HNSCC): cellular mechanisms of radioresistance and clinical relevance. Cancer Lett. 386, 87–99 (2017).
Puck, T. T., Cieciura, S. J. & Fisher, H. W. Clonal growth in vitro of human cells with fibroblastic morphology; comparison of growth and genetic characteristics of single epithelioid and fibroblast-like cells from a variety of human organs. J. Exp. Med. 106, 145–158 (1957).
Fisher, H. W. & Puck, T. T. On the functions of x-irradiated “feeder” cells in supporting growth of single mammalian cells. Proc. Natl Acad. Sci. USA 42, 900–906 (1956).
Terasima, T. & Tolmach, L. J. Changes in x-ray sensitivity of HeLa cells during the division cycle. Nature 190, 1210–1211 (1961).
Son, J. et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013).
Varfolomeev, E. et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131, 669–681 (2007).
Till, J. E. & McCulloch, C. E. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 14, 213–222 (1961).
Becker, A. J., McCulloch, C. E. & Till, J. E. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature 197, 452–454 (1963).
Park, C. H., Bergsagel, D. E. & McCulloch, E. A. Mouse myeloma tumor stem cells: a primary cell culture assay. J. Natl Cancer Inst. 46, 411–422 (1971).
Courtenay, V. D. & Mills, J. An in vitro colony assay for human tumours grown in immune-suppressed mice and treated in vivo with cytotoxic agents. Br. J. Cancer 37, 261–268 (1978).
Hamburger, A. W. & Salmon, S. E. Primary bioassay of human tumor stem cells. Science 197, 461–463 (1977).
Whitlock, C. A. & Witte, O. N. Long-term culture of B lymphocytes and their precursors from murine bone marrow. Proc. Natl Acad. Sci. USA 79, 3608–3612 (1982).
Kondo, M., Weissman, I. L. & Akashi, K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91, 661–672 (1997).
Matsui, W. et al. Characterization of clonogenic multiple myeloma cells. Blood 103, 2332–2336 (2004).
Eliason, J. F., Aapro, M. S., Decrey, D. & Brink-Petersen, M. Non-linearity of colony formation by human tumour cells from biopsy samples. Br. J. Cancer 52, 311–318 (1985).
Pomp, J. et al. Cell density dependent plating efficiency affects outcome and interpretation of colony forming assays. Radiother. Oncol. 40, 121–125 (1996).
Veldwijk, M. R., Zhang, B., Wenz, F. & Herskind, C. The biological effect of large single doses: a possible role for non-targeted effects in cell inactivation. PLoS One 9, e84991 (2014).
Adrian, G., Ceberg, C., Carneiro, A. & Ekblad, L. Rescue effect inherited in colony formation assays affects radiation response. Radiat. Res. 189, 44–52 (2018).
Dakhore, S., Nayer, B. & Hasegawa, K. Human pluripotent stem cell culture: current status, challenges, and advancement. Stem Cells Int. 2018, 7396905 (2018).
Repetto, G., del Peso, A. & Zurita, J. L. Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat. Protoc. 3, 1125–1131 (2008).
Lindhagen, E., Nygren, P. & Larsson, R. The fluorometric microculture cytotoxicity assay. Nat. Protoc. 3, 1364–1369 (2008).
Riss, T. L. et al. Cell viability assays. in Assay Guidance Manual (eds. Markossian S. et al.) (Eli Lilly and the National Center for Advancing Translational Sciences, 2004).
Rampersad, S. N. Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays. Sensors (Basel) 12, 12347–12360 (2012).
Lundin, A., Hasenson, M., Persson, J. & Pousette, A. Estimation of biomass in growing cell lines by adenosine triphosphate assay. Methods Enzymol. 133, 27–42 (1986).
Temple, S. Division and differentiation of isolated CNS blast cells in microculture. Nature 340, 471–473 (1989).
Davis, A. A. & Temple, S. A self-renewing multipotential stem cell in embryonic rat cerebral cortex. Nature 372, 263–266 (1994).
Watanabe, K. et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 25, 681–686 (2007).
Ploemacher, R. E., van der Sluijs, J. P., Voerman, J. S. & Brons, N. H. An in vitro limiting-dilution assay of long-term repopulating hematopoietic stem cells in the mouse. Blood 74, 2755–2763 (1989).
Sutherland, H. J., Lansdorp, P. M., Henkelman, D. H., Eaves, A. C. & Eaves, C. J. Functional characterization of individual human hematopoietic stem cells cultured at limiting dilution on supportive marrow stromal layers. Proc. Natl Acad. Sci. USA 87, 3584–3588 (1990).
Fogg, D. K. et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83–87 (2006).
Riether, C. et al. Targeting CD70 with cusatuzumab eliminates acute myeloid leukemia stem cells in patients treated with hypomethylating agents. Nat. Med. 26, 1459–1467 (2020).
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009).
Fiebig, H. H., Maier, A. & Burger, A. M. Clonogenic assay with established human tumour xenografts: correlation of in vitro to in vivo activity as a basis for anticancer drug discovery. Eur. J. Cancer 40, 802–820 (2004).
Borowicz, S. et al. The soft agar colony formation assay. J. Vis. Exp. 92, e51998 (2014).
Leivas, A. et al. Natural killer cells efficiently target multiple myeloma clonogenic tumor cells. Cancer Immunol. Immunother. (in the press).
Kukreja, A. et al. Enhancement of clonogenicity of human multiple myeloma by dendritic cells. J. Exp. Med. 203, 1859–1865 (2006).
Eke, I., Hehlgans, S., Sandfort, V. & Cordes, N. 3D matrix-based cell cultures: automated analysis of tumor cell survival and proliferation. Int. J. Oncol 48, 313–321 (2016).
Nielson, L., Smyth, G. & Greenfield, P. Hemacytometer cell count distributions: implications of non-Poisson behavior. Biotechnol. Prog. 7, 560–563 (1991).
Harnicek, D. et al. Hyperthermia adds to trabectedin effectiveness and thermal enhancement is associated with BRCA2 degradation and impairment of DNA homologous recombination repair. Int. J. Cancer 139, 467–479 (2016).
Weiss, E. M. et al. High hydrostatic pressure treatment generates inactivated mammalian tumor cells with immunogeneic features. J. Immunotoxicol. 7, 194–204 (2010).
Preciado, S. et al. The incorporation of extracellular vesicles from mesenchymal stromal cells into CD34+ cells increases their clonogenic capacity and bone marrow lodging ability. Stem Cells 37, 1357–1368 (2019).
Yao, T. & Asayama, Y. Animal-cell culture media: history, characteristics, and current issues. Reprod. Med. Biol. 16, 99–117 (2017).
Nuryadi, E., Mayang Permata, T. B., Komatsu, S., Oike, T. & Nakano, T. Inter-assay precision of clonogenic assays for radiosensitivity in cancer cell line A549. Oncotarget 9, 13706–13712 (2018).
Lacerda, L. et al. Simvastatin radiosensitizes differentiated and stem-like breast cancer cell lines and is associated with improved local control in inflammatory breast cancer patients treated with postmastectomy radiation. Stem Cells Transl. Med. 3, 849–856 (2014).
Dahle, J., Kakar, M., Steen, H. B. & Kaalhus, O. Automated counting of mammalian cell colonies by means of a flat bed scanner and image processing. Cytometry A 60, 182–188 (2004).
Choudhry, P. High-throughput method for automated colony and cell counting by digital image analysis based on edge detection. PLoS One 11, e0148469 (2016).
Bewes, J. M., Suchowerska, N. & McKenzie, D. R. Automated cell colony counting and analysis using the circular Hough image transform algorithm (CHiTA). Phys. Med. Biol. 53, 5991–6008 (2008).
Meijering, E., Carpenter, A. E., Peng, H., Hamprecht, F. A. & Olivo-Marin, J. C. Imagining the future of bioimage analysis. Nat. Biotechnol. 34, 1250–1255 (2016).
Moen, E. et al. Deep learning for cellular image analysis. Nat. Methods 16, 1233–1246 (2019).
Segebarth, D. et al. On the objectivity, reliability, and validity of deep learning enabled bioimage analyses. eLife 9, e59780 (2020).
Parker, W. S. & Risbey, J. S. False precision, surprise and improved uncertainty assessment. Philos. Trans. A Math. Phys. Eng. Sci. 373, 20140453 (2015).
Kellerer, A. M. & Rossi, H. H. A generalized formulation of dual radiation action. Radiat. Res. 75, 471–488 (1978).
Unkel, S., Belka, C. & Lauber, K. On the analysis of clonogenic survival data: statistical alternatives to the linear-quadratic model. Radiat. Oncol. 11, 11 (2016).
Vembadi, A., Menachery, A. & Qasaimeh, M. A. Cell cytometry: review and perspective on biotechnological advances. Front. Bioeng. Biotechnol. 7, 147 (2019).
Gerweck, L. E., Dullea, R., Zaidi, S. T., Budach, W. & Hartford, A. Influence of experimental factors on intrinsic radiosensitivity assays at low doses of radiation: cell multiplicity. Radiat. Res. 138, 361–366 (1994).
Rockwell, S. Effects of clumps and clusters on survival measurements with clonogenic assays. Cancer Res. 45, 1601–1607 (1985).
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft DFG (SFB1321, Project-ID 329628492, P16), the Bundesministerium fuer Bildung und Forschung BMBF (ZiSStrans NUK047A, NUK047C, METABOLiST NUK061C and DKTK) and the International graduate program iTarget (Elitenetzwerk Bayern).
Author information
Authors and Affiliations
Contributions
N.B. and K.L. conceived and designed the protocol with support from D.S., H.Z. and C.B. N.B. acquired the data. D.S. provided mathematical consultation and developed and programmed the R-package CFAcoop. The MS Excel template files for power regression–based and PE-based survival analysis were generated by N.B. and D.S. N.B. and K.L. wrote the manuscript, and all authors commented on and discussed the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Tatsuya Ohno, Daniel Wahl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Related links
Key references using this protocol
Brix, N. et al. Radiat. Oncol. 15, 248 (2020): https://doi.org/10.1186/s13014-020-01697-y
Orth, M. et al. Oncogene 37, 52 (2018): https://doi.org/10.1038/onc.2017.304
Hess, J. et al. Cancer Lett. 386, 87 (2017): https://doi.org/10.1016/j.canlet.2016.11.014
This protocol is an extension to: Nat. Protoc. 1, 2315–2319 (2006): https://doi.org/10.1038/nprot.2006.339
Supplementary information
Supplementary Table 1
MS Excel template file for power regression–based analysis of clonogenic survival
Supplementary Table 2
MS Excel template file for PE-based analysis of clonogenic survival
Source data
Source Data Fig. 5
All colony-counting raw data
Source Data Fig. 9
All colony-counting raw data
Rights and permissions
About this article
Cite this article
Brix, N., Samaga, D., Belka, C. et al. Analysis of clonogenic growth in vitro. Nat Protoc 16, 4963–4991 (2021). https://doi.org/10.1038/s41596-021-00615-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00615-0
This article is cited by
-
Combining a noble gas with radiotherapy: glutamate receptor antagonist xenon may act as a radiosensitizer in glioblastoma
Radiation Oncology (2024)
-
PLK4 as a potential target to enhance radiosensitivity in triple-negative breast cancer
Radiation Oncology (2024)
-
Pan-cancer ion transport signature reveals functional regulators of glioblastoma aggression
The EMBO Journal (2024)
-
Systematic in vitro analysis of therapy resistance in glioblastoma cell lines by integration of clonogenic survival data with multi-level molecular data
Radiation Oncology (2023)
-
IL-1RA promotes oral squamous cell carcinoma malignancy through mitochondrial metabolism-mediated EGFR/JNK/SOX2 pathway
Journal of Translational Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.