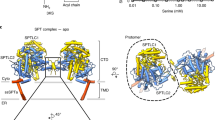
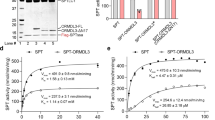
Abstract
Human serine palmitoyltransferase (SPT) complex catalyzes the initial and rate-limiting step in the de novo biosynthesis of all sphingolipids. ORMDLs regulate SPT function, with human ORMDL3 being related to asthma. Here we report three high-resolution cryo-EM structures: the human SPT complex, composed of SPTLC1, SPTLC2 and SPTssa; the SPT–ORMDL3 complex; and the SPT–ORMDL3 complex bound to two substrates, PLP-l-serine (PLS) and a non-reactive palmitoyl-CoA analogue. SPTLC1 and SPTLC2 form a dimer of heterodimers as the catalytic core. SPTssa participates in acyl-CoA coordination, thereby stimulating the SPT activity and regulating the substrate selectivity. ORMDL3 is located in the center of the complex, serving to stabilize the SPT assembly. Our structural and biochemical analyses provide a molecular basis for the assembly and substrate selectivity of the SPT and SPT–ORMDL3 complexes, and lay a foundation for mechanistic understanding of sphingolipid homeostasis and for related therapeutic drug development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
EM density maps have been deposited in the EMDB under the accession codes EMD-30081 (SPT, dimer), EMD-30082 (SPT, monomer), EMD-30079 (SPT–ORMDL3, dimer), EMD-30080 (SPT–ORMDL3, monomer), EMD-30441 (substrate-bound SPT–ORMDL3, monomer, class I) and EMD-30442 (substrate-bound SPT–ORMDL3, monomer, class II). Atomic coordinates have been deposited in the PDB under the accession codes PDB 6M4N (SPT–ORMDL3, dimer), PDB 6M4O (SPT–ORMDL3, monomer), PDB 7CQI (substrate-bound SPT–ORMDL3, monomer, class I) and PDB 7CQK (substrate-bound SPT–ORMDL3, monomer, class II). Source data are available with the paper online. Source data are provided with this paper.
References
Harayama, T. & Riezman, H. Understanding the diversity of membrane lipid composition. Nat. Rev. Mol. Cell Biol. 19, 281–296 (2018).
Hannun, Y. A. & Obeid, L. M. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150 (2008).
Merrill, A. H. Jr. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 111, 6387–6422 (2011).
Hanada, K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta 1632, 16–30 (2003).
Lowther, J., Naismith, J. H., Dunn, T. M. & Campopiano, D. J. Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem. Soc. Trans. 40, 547–554 (2012).
Harrison, P. J., Dunn, T. M. & Campopiano, D. J. Sphingolipid biosynthesis in man and microbes. Nat. Prod. Rep. 35, 921–954 (2018).
Nagiec, M. M., Lester, R. L. & Dickson, R. C. Sphingolipid synthesis: identification and characterization of mammalian cDNAs encoding the Lcb2 subunit of serine palmitoyltransferase. Gene 177, 237–241 (1996).
Weiss, B. & Stoffel, W. Human and murine serine-palmitoyl-CoA transferase—cloning, expression and characterization of the key enzyme in sphingolipid synthesis. Eur. J. Biochem. 249, 239–247 (1997).
Hanada, K. et al. A mammalian homolog of the yeast LCB1 encodes a component of serine palmitoyltransferase, the enzyme catalyzing the first step in sphingolipid synthesis. J. Biol. Chem. 272, 32108–32114 (1997).
Hornemann, T., Richard, S., Rutti, M. F., Wei, Y. & von Eckardstein, A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J. Biol. Chem. 281, 37275–37281 (2006).
Bejaoui, K. et al. SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat. Genet. 27, 261–262 (2001).
Dawkins, J. L., Hulme, D. J., Brahmbhatt, S. B., Auer-Grumbach, M. & Nicholson, G. A. Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat. Genet. 27, 309–312 (2001).
Rotthier, A. et al. Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. Am. J. Hum. Genet. 87, 513–522 (2010).
Mandon, E. C., van Echten, G., Birk, R., Schmidt, R. R. & Sandhoff, K. Sphingolipid biosynthesis in cultured neurons. Eur. J. Biochem. 198, 667–674 (1991).
Hornemann, T., Wei, Y. & von Eckardstein, A. Is the mammalian serine palmitoyltransferase a high-molecular-mass complex? Biochemical J. 405, 157–164 (2007).
Han, G. et al. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl Acad. Sci. USA 106, 8186–8191 (2009).
Hornemann, T. et al. The SPTLC3 subunit of serine palmitoyltransferase generates short chain sphingoid bases. J. Biol. Chem. 284, 26322–26330 (2009).
Harmon, J. M. et al. Topological and functional characterization of the ssSPTs, small activating subunits of serine palmitoyltransferase. J. Biol. Chem. 288, 10144–10153 (2013).
Russo, S. B., Tidhar, R., Futerman, A. H. & Cowart, L. A. Myristate-derived d16:0 sphingolipids constitute a cardiac sphingolipid pool with distinct synthetic routes and functional properties. J. Biol. Chem. 288, 13397–13409 (2013).
Zhao, L. et al. Elevation of 20-carbon long chain bases due to a mutation in serine palmitoyltransferase small subunit b results in neurodegeneration. Proc. Natl Acad. Sci. USA 112, 12962–12967 (2015).
Breslow, D. K. et al. Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–U1065 (2010).
Han, S. M., Lone, M. A., Schneiter, R. & Chang, A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl Acad. Sci. USA 107, 5851–5856 (2010).
Davis, D., Kannan, M. & Wattenberg, B. Orm/ORMDL proteins: Gate guardians and master regulators. Adv. Biol. Regul. 70, 3–18 (2018).
Hjelmqvist, L. et al. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 3, Research0027 (2002).
Moffatt, M. F. et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 (2007).
Moffatt, M. F. et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363, 1211–1221 (2010).
Yard, B. A. et al. The structure of serine palmitoyltransferase; gateway to sphingolipid biosynthesis. J. Mol. Biol. 370, 870–886 (2007).
Raman, M. C. et al. The external aldimine form of serine palmitoyltransferase: structural, kinetic, and spectroscopic analysis of the wild-type enzyme and HSAN1 mutant mimics. J. Biol. Chem. 284, 17328–17339 (2009).
Ikushiro, H. et al. Structural insights into the enzymatic mechanism of serine palmitoyltransferase from Sphingobacterium multivorum. J. Biochem. 146, 549–562 (2009).
Raman, M. C., Johnson, K. A., Clarke, D. J., Naismith, J. H. & Campopiano, D. J. The serine palmitoyltransferase from Sphingomonas wittichii RW1: An interesting link to an unusual acyl carrier protein. Biopolymers 93, 811–822 (2010).
Harrison, P. J. et al. Use of isotopically labeled substrates reveals kinetic differences between human and bacterial serine palmitoyltransferase. J. Lipid Res. 60, 953–962 (2019).
Hanada, K., Hara, T. & Nishijima, M. Purification of the serine palmitoyltransferase complex responsible for sphingoid base synthesis by using affinity peptide chromatography techniques. J. Biol. Chem. 275, 8409–8415 (2000).
Ikushiro, H., Fujii, S., Shiraiwa, Y. & Hayashi, H. Acceleration of the substrate Calpha deprotonation by an analogue of the second substrate palmitoyl-CoA in serine palmitoyltransferase. J. Biol. Chem. 283, 7542–7553 (2008).
Cantalupo, A. et al. Nogo-B regulates endothelial sphingolipid homeostasis to control vascular function and blood pressure. Nat. Med. 21, 1028–1037 (2015).
Su, W. C., Lin, Y. H., Pagac, M. & Wang, C. W. Seipin negatively regulates sphingolipid production at the ER-LD contact site. J. Cell Biol. 218, 3663–3680 (2019).
Siow, D. L. & Wattenberg, B. W. Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J. Biol. Chem. 287, 40198–40204 (2012).
Siow, D., Sunkara, M., Dunn, T. M., Morris, A. J. & Wattenberg, B. ORMDL/serine palmitoyltransferase stoichiometry determines effects of ORMDL3 expression on sphingolipid biosynthesis. J. Lipid Res. 56, 898–908 (2015).
Davis, D. L., Gable, K., Suemitsu, J., Dunn, T. M. & Wattenberg, B. W. The ORMDL/Orm-serine palmitoyltransferase (SPT) complex is directly regulated by ceramide: Reconstitution of SPT regulation in isolated membranes. J. Biol. Chem. 294, 5146–5156 (2019).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Grigorieff, N. & Grant, T. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4, e06980 (2015).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, https://doi.org/10.7554/eLife.42166 (2018).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Chen, S. X. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 66, 213–221 (2010).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
Acknowledgements
We thank the cryo-EM facilities of Southern University of Science and Technology and Princeton Imaging and Analysis Center for providing the facility support. We thank H. Ikushiro (Osaka Medical College, Japan) for providing the S-CoA. This work was supported by the National Natural Science Foundation of China (92057101, X.G.), the Natural Science Foundation of Guangdong Province of China for Distinguished Young Scientists (2019B151502047, X.G.) and the Guangdong Innovation Research Team Fund (2016ZT06S172, S.L.).
Author information
Authors and Affiliations
Contributions
X.G. conceived and supervised the project. X.G., S.L and T.X. designed experiments. X.G., S.L., T.X., P.L. and L.W. performed the experiments. All authors contributed to data analysis. S.L. and T.X. contributed to manuscript preparation. X.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Structural & Molecular Biology thanks Binks Wattenberg, Ming Zhou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Florian Ullrich and Anke Sparmann were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 In vitro functional characterization and cryo-EM analysis of the SPT complex.
a, Size exclusion chromatography (SEC) profile and Coomassie blue−stained SDS-PAGE gel corresponding to the last SPT complex purification step. The peak fractions indicated by the arrow were pooled and concentrated for biochemical and structural characterization. b, Initial rate of reaction versus L-serine concentration measured using the purified wild-type (WT) SPT complex or catalytic mutant (K379A in SPTLC2) SPT complex. Each data point is the average of three independent experiments, and error bars represent the SEM. The data points were fitted with a Michaelis-Menten equation. c, Initial rate of reaction versus palmitoyl-CoA concentration for the WT SPT complex. The data points were fitted with an allosteric sigmoidal equation. Please refer to the Methods section for details of the SPT activity assay. d, Representative cryo-EM micrograph and 2D class averages of the SPT complex. e, Flowchart for cryo-EM data processing. f, Angular distribution of the particles used to reconstruct the dimeric SPT complex. g, Gold-standard FSC curves for the dimeric (black curve) and monomeric (purple curve) SPT complexes generated using Relion 3.0. h, Local resolution maps of the dimeric (left) and monomeric (right) SPT complexes. The color code for resolution, shown with the unit Å, was calculated using Relion 3.0. Uncropped image for a and data for graphs b,c are available as source data.
Extended Data Fig. 2 Cryo-EM analysis of the SPT–ORMDL3 complex.
a, Representative cryo-EM micrograph (left) and 2D class averages (right) of the SPT–ORMDL3 complex. b, Flowchart for cryo-EM data processing. c, Angular distribution of the particles used to reconstruct the dimeric SPT–ORMDL3 complex. d, Gold-standard FSC curves for the dimeric (black curve) and monomeric (purple curve) SPT–ORMDL3 complexes. e, Local resolution maps of the dimeric (left) and monomeric (right) SPT–ORMDL3 complexes. f, Directional FSC (dFSC) to assess directional isotropy for the monomeric SPT–ORMDL3 map. g, Calculated FSC curves between the refined model and the half map used for refinement (blue), the other half map (green), and the full map (red).
Extended Data Fig. 3 EM maps of the SPT–ORMDL3 complex.
a, 3D EM reconstruction of the dimeric SPT–ORMDL3 complex, with density covering individual subunits colored same as in Fig. 1e. The detergent micelles, displayed at a low contour level, are shown as gray, semi-transparent surface to indicate the orientation of transmembrane domain. b,c, EM maps corresponding to representative segments (b) and active/nonfunctional sites (c) of the monomeric substrate−free SPT–ORMDL3 complex. Electron density maps contoured at 5 σ were prepared in PyMol.
Extended Data Fig. 4 EM reconstruction of the SPT complex.
a, The 3D EM reconstruction of the SPT complex shown from two different views. The subunits are colored in the same manner used to color the SPT–ORMDL3 complex in Fig. 1e. b, Structural comparison of the SPT and SPT–ORMDL3 complexes. The EM density for monomeric SPT complex could be well fitted with the corresponding structural model from the SPT–ORMDL3 complex.
Extended Data Fig. 5 The dimerization interface in the SPT–ORMDL3 complex.
a, Structure of a dimer of SPTLC1–SPTLC2 heterodimers. The dimer-dimer interface contains only limited interactions in the cytosol, which mainly involve the small helix α1 of SPTLC1 and the α6-β4 loop of SPTLC2. b, Biochemical validation of the structurally revealed dimer-dimer interface. The Mut2T variant of the SPT complex eluted around 1.5 ml later than the WT SPT complex in SEC. c, Representative cryo-EM micrograph and 2D class averages of the SPT Mut2T variant. d, The SPT Mut2T variant showed enzymatic activity comparable to that of the WT SPT complex when challenged with L-serine and palmitoyl-CoA. Uncropped image for b and data for graph d are available as source data.
Extended Data Fig. 6 Structural comparison of SPTLC1–SPTLC2 heterodimer and bacterial spSPT homodimer shown from two different views.
Superimposition of the spSPT homodimer (PDB 2JG2) onto the SPTLC1–SPTLC2 heterodimer by Cα atoms. Except for the N-terminal extensions of SPTLC1 and SPTLC2, the two structures are highly similar: superimposition of the two structures over 586 aligned Cα atoms yielded an RMSD of 1.5 Å.
Extended Data Fig. 7 Cryo-EM analysis of the substrate-bound SPT–ORMDL3 complex.
a, Chemical structures of palmitoyl-CoA and S-(2-oxoheptadecyl)-CoA (S-CoA). The site of -CH2- group insertion in the S-CoA is indicated by a red arrow. b, Representative cryo-EM micrograph (left) and 2D class averages (right) of substrate-bound SPT–ORMDL3. c, Flowchart for cryo-EM data processing. d, Gold-standard FSC curves for the two monomeric reconstructions. e, Local resolution maps of the two monomeric substrate-bound reconstructions.
Extended Data Fig. 8 EM maps corresponding to representative segments (a) and substrate binding site (b) of the monomeric substrate-bound SPT–ORMDL3 complex (class I).
The map for ORMDL3-TM3 is contoured at 3σ, and all the other maps are contoured at 5 σ.
Extended Data Fig. 9 Purification of SPT–ORMDL3 mutants related to substrate binding and the substrate preferences of WT SPT and SPT–ORMDL3 complexes.
a, Purification of SPT–ORMDL3 complexes containing mutations related to PLS coordination. b, Relative catalytic activity of the WT SPT and SPT–ORMDL3 complexes against three different amino acid substrates. c, Purification of SPT–ORMDL3 complexes containing mutations related to S-CoA coordination. d, Relative catalytic activity of the WT SPT and SPT–ORMDL3 complexes against three different acyl-CoA substrates. e, Purification of SPT–ORMDL3 complexes containing SPTssaM28V or SPTssaM28G. Uncropped images for a, c, e and date for graphs b, d are available as source data.
Extended Data Fig. 10 Enzymatic kinetics of several SPT–ORMDL3 mutants and the effects of C6-ceramide on the enzymatic activities of purified SPT and SPT–ORMDL3 complexes.
a, Enzymatic kinetics of two SPT–ORMDL3 mutants related to PLS coordination. b, Enzymatic kinetics of two SPT–ORMDL3 mutants related to S-CoA coordination. c, C6-ceramide has little effect on the enzymatic activities for both SPT and SPT–ORMDL3 complexes. The SPT and SPT–ORMDL3 complexes were incubated with 30 μM DMSO-solubilized C6-ceramide before the enzymatic activities were measured. Date for graphs a-c are available as source data.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed gel images.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed gel images.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed gel images.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed gel images.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Li, S., Xie, T., Liu, P. et al. Structural insights into the assembly and substrate selectivity of human SPT–ORMDL3 complex. Nat Struct Mol Biol 28, 249–257 (2021). https://doi.org/10.1038/s41594-020-00553-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-020-00553-7
This article is cited by
-
Sphingolipids: drivers of cardiac fibrosis and atrial fibrillation
Journal of Molecular Medicine (2024)
-
Structure of the ceramide-bound SPOTS complex
Nature Communications (2023)
-
Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis
Nature Communications (2023)
-
Convergent evolution of bacterial ceramide synthesis
Nature Chemical Biology (2022)
-
Contribution of specific ceramides to obesity-associated metabolic diseases
Cellular and Molecular Life Sciences (2022)