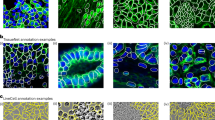
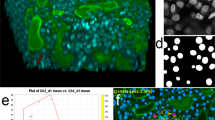
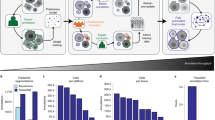
Abstract
Many biological applications require the segmentation of cell bodies, membranes and nuclei from microscopy images. Deep learning has enabled great progress on this problem, but current methods are specialized for images that have large training datasets. Here we introduce a generalist, deep learning-based segmentation method called Cellpose, which can precisely segment cells from a wide range of image types and does not require model retraining or parameter adjustments. Cellpose was trained on a new dataset of highly varied images of cells, containing over 70,000 segmented objects. We also demonstrate a three-dimensional (3D) extension of Cellpose that reuses the two-dimensional (2D) model and does not require 3D-labeled data. To support community contributions to the training data, we developed software for manual labeling and for curation of the automated results. Periodically retraining the model on the community-contributed data will ensure that Cellpose improves constantly.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The manually segmented cytoplasmic dataset is available at www.cellpose.org/dataset and https://doi.org/10.25378/janelia.13270466.
Code availability
The reviewed version of Cellpose is available as Supplementary Software. The code, graphical user interface and updated versions are available at www.github.com/mouseland/cellpose. To test out the model directly on the web, visit www.cellpose.org. Note that the test time augmentations and tiling, which improve segmentation, are not performed on the website to save computation time.
References
Boutros, M., Heigwer, F. & Laufer, C. Microscopy-based high-content screening. Cell 163, 1314–1325 (2015).
Sommer, C., Straehle, C., Koethe, U. & Hamprecht, F. A. Ilastik: interactive learning and segmentation toolkit. IEEE International Symposium on Biomedical Imaging, 230–233 (2011).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation. Preprint at arXiv 1505.04597 (2015).
Apthorpe, N. et al. Automatic neuron detection in calcium imaging data using convolutional networks. Advances in Neural Information Processing Systems 29, 3270–3278 (2016).
Guerrero-Pena, F. A. et al. Multiclass weighted loss for instance segmentation of cluttered cells. In Proc. 2018 25th IEEE International Conference on Image Processing (ICIP) 2451–2455 (IEEE, 2018).
Xie, W., Noble, J. A. & Zisserman, A. Microscopy cell counting and detection with fully convolutional regression networks. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 6, 283–292 (2018).
Al-Kofahi, Y., Zaltsman, A., Graves, R., Marshall, W. & Rusu, M. A deep learning-based algorithm for 2-D cell segmentation in microscopy images. BMC Bioinformatics 19, 1–11 (2018).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
McQuin, C. et al. Cellprofiler 3.0: next-generation image processing for biology. PLoS Biology 16, e2005970 (2018).
Carpenter, A. E. et al. Cellprofiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Chen, J. et al. The Allen cell structure segmenter: a new open source toolkit for segmenting 3D intracellular structures in fluorescence microscopy images. Preprint at https://www.biorxiv.org/content/10.1101/491035v1 (2018).
Funke, J., Mais, L., Champion, A., Dye, N. & Kainmueller, D. A benchmark for epithelial cell tracking. Proceedings of the European Conference on Computer Vision https://doi.org/10.1007/978-3-030-11024-6_33 (2018).
Yi, J. et al. Object-guided instance segmentation for biological images. Preprint at https://arxiv.org/abs/1911.09199 (2019).
Caicedo, J. C. et al. Nucleus segmentation across imaging experiments: the 2018 data science bowl. Nat. Methods 16, 1247–1253 (2019).
He, K., Gkioxari, G., Dollár, P. & Girshick, R. Mask R-CNN. Preprint at https://arxiv.org/abs/1703.06870 (2018).
Abdulla, W. Mask r-cnn for object detection and instance segmentation on keras and tensorflow (GitHub, 2017); https://github.com/matterport/Mask_RCNN
Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. Cell detection with star-convex polygons. International Conference on Medical Image Computing and Computer-Assisted Intervention 265–273 (2018).
Hollandi, R. et al. A deep learning framework for nucleus segmentation using image style transfer. Preprint at https://www.biorxiv.org/content/10.1101/580605v1 (2019).
Van Rossum, G. & Drake, F. L. Python 3 Reference Manual (CreateSpace, 2009).
Harris, C. R. et al. Array programming with numpy. Nature 585, 357–362 (2020).
Jones, E. et al. SciPy: open source scientific tools for Python (SciPy, 2001); http://www.scipy.org/
Lam, S. K., Pitrou, A. & Seibert, S. Numba: a llvm-based python jit compiler. In Proc. Second Workshop on the LLVM Compiler Infrastructure in HPC 7 (2015); https://doi.org/10.1145/2833157.2833162
Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools 25, 120–125 (2000).
Chen, T. et al. Mxnet: A flexible and efficient machine learning library for heterogeneous distributed systems. Preprint at https://arxiv.org/abs/1512.01274 (2015).
Summerfield, M. Rapid GUI Programming with Python and Qt: The Definitive Guide to PyQt Programming (Pearson Education, 2007).
Campagnola, L. Scientific graphics and GUI library for Python; http://pyqtgraph.org/
Beucher, S. & Meyer, F. in Mathematical Morphology in Image Processing (ed. Dougherty, E. R.) 433–481 (Marcel Dekker, 1993).
Li, G. et al. Segmentation of touching cell nuclei using gradient flow tracking. J. Microsc. 231, 47–58 (2008).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition 770–778 (2016).
Gatys, L. A., Ecker, A. S. & Bethge, M. Image style transfer using convolutional neural networks. Proceedings of the IEEE conference on computer vision and pattern recognition 2414–2423 (2016).
Karras, T., Laine, S. & Aila, T. A style-based generator architecture for generative adversarial networks. Proceedings of the IEEE conference on computer vision and pattern recognition 4401–4410 (2019).
OMERO. Image data resource (IDR, 2020); https://idr.openmicroscopy.org/cell/
Williams, E. et al. Image data resource: a bioimage data integration and publication platform. Nat. Methods 14, 775–781 (2017).
Maaten, Lvd & Hinton, G. Visualizing data using t-SNE. J. Machine Learning Res. 9, 2579–2605 (2008).
Pedregosa, F. et al. Scikit-learn: Machine learning in python. J. Machine Learning Res. 12, 2825–2830 (2011).
Yu, W., Lee, H. K., Hariharan, S., Bu, W. Y. & Ahmed, S. Ccdb:6843, Mus musculus, neuroblastoma. Cell Image Library https://doi.org/10.7295/W9CCDB6843
Bai, X.-c, McMullan, G. & Scheres, S. H. How cryo-em is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 (2015).
Weigert, M., Schmidt, U., Haase, R., Sugawara, K. & Myers, G. Star-convex polyhedra for 3D object detection and segmentation in microscopy. Preprint at https://arxiv.org/abs/1908.03636 (2019).
Ulman, V. et al. An objective comparison of cell-tracking algorithms. Nat. Methods 14, 1141–1152 (2017).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Arbelaez, P., Maire, M., Fowlkes, C. & Malik, J. Contour detection and hierarchical image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 33, 898–916 (2010).
Kumar, N. et al. A dataset and a technique for generalized nuclear segmentation for computational pathology. IEEE Trans. Med. Imag. 36, 1550–1560 (2017).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Kluyver, T. et al. Jupyter notebooks-a publishing format for reproducible computational workflows. ELPUB 87–90 (2016).
Ljosa, V., Sokolnicki, K. L. & Carpenter, A. E. Annotated high-throughput microscopy image sets for validation. Nat. Methods 9, 637–637 (2012).
Jones, T. R., Carpenter, A. & Golland, P. Voronoi-based segmentation of cells on image manifolds. International Workshop on Computer Vision for Biomedical Image Applications 535–543 (2005).
Rapoport, D. H., Becker, T., Mamlouk, A. M., Schicktanz, S. & Kruse, C. A novel validation algorithm allows for automated cell tracking and the extraction of biologically meaningful parameters. PLoS ONE 6, e27315 (2011).
Raza, S. E. A. et al. Micro-Net: a unified model for segmentation of various objects in microscopy images. Med. Image Anal. 52, 160–173 (2019).
Lopuhin, K. kaggle-dsbowl-2018-dataset-fixes (GitHub, 2018); https://github.com/lopuhin/kaggle-dsbowl-2018-dataset-fixes
Kumar, N. et al. A multi-organ nucleus segmentation challenge. IEEE Trans. Med. Imag. 39, 1380–1391 (2019).
Coelho, L. P., Shariff, A. & Murphy, R. F. Nuclear segmentation in microscope cell images: a hand-segmented dataset and comparison of algorithms. IEEE International Symposium on Biomedical Imaging: From Nano to Macro 518–521 (2009).
Chen, F., Tillberg, P. W. & Boyden, E. S. Expansion microscopy. Science 347, 543–548 (2015).
Chen, F. et al. Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13, 679–684 (2016).
Cao, Z., Hidalgo, G., Simon, T., Wei, S.-E. & Sheikh, Y. OpenPose: realtime multi-person 2D pose estimation using part affinity fields. Preprint at arXiv 1812.08008 (2019).
Neven, D., Brabandere, B. D., Proesmans, M. & Gool, L. V. Instance segmentation by jointly optimizing spatial embeddings and clustering bandwidth. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 8837–8845 (2019).
Gorelick, L., Galun, M., Sharon, E., Basri, R. & Brandt, A. Shape representation and classification using the poisson equation. IEEE Trans. Pattern Anal. Mach. Intell. 28, 1991–2005 (2006).
He, K., Zhang, X., Ren, S. & Sun, J. Identity mappings in deep residual networks. Preprint at arXiv 1603.05027 (2016).
Chaurasia, A. & Culurciello, E. LinkNet: exploiting encoder representations for efficient semantic segmentation. In Proc. 2017 IEEE Visual Communications and Image Processing (VCIP) 1–4 (IEEE, 2017).
Schmidt, U. & Weigert, M. StarDist—object detection with star-convex shapes (GitHub, 2019); http://github.com/mpicbg-csbd/stardist
Mingqiang, Y., Kidiyo, K. & Joseph, R. A survey of shape feature extraction techniques. Pattern Recog. 15, 43–90 (2008).
Acknowledgements
This research was funded by the Howard Hughes Medical Institute at the Janelia Research Campus. We thank P. Tillberg and members of the Tillberg laboratory for advice related to expansion microscopy, and W. Sun for sharing calcium imaging data from mouse hippocampus. We also thank S. Saalfeld and J. Funke for useful discussions.
Author information
Authors and Affiliations
Contributions
C.S. and M.P. designed the study. T.W. acquired data. M.M. manually segmented images. C.S., M.P. and T.W. performed data analysis. C.S. and M.P. wrote the manuscript, with feedback from T.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Rita Strack was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Graphical User Interface (GUI).
Shown in the GUI is an image from the test set, zoomed in on an area of interest, and segmented using Cellpose. The GUI serves two main purposes: 1) easily run Cellpose ‘out-of-the-box’ on new images and visualize the results in an interactive mode; 2) manually segment new images, to provide training data for Cellpose. The image view can be changed between image channels, cellpose vector flows and cellpose predicted pixel probabilities. Similarly, the mask overlays can be changed between outlines, transparent masks or both. The size calibration procedure can be run to estimate cell size, or the user can directly input the cell diameter, with an image overlay of a red disk shown as an aid for visual calibration. Dense, complete manual segmentations can be uploaded to our server with one button press, and the latest trained model can be downloaded from the drop-down menu.
Extended Data Fig. 2 Effect of network architecture and training set exclusions.
a-d, Each analysis compares average precision relative to the standard 4-network average, full Cellpose architecture. The following changes were made: a, removed the style vectors; b, replaced residual blocks with standard convolutional layers; c, Replaced addition with concatenation on the upsampling pass; d, standard Unet architecture (in other words a, b and c combined). e, Performance on generalized data for the ‘generalist’ Cellpose model, and a version of the model trained without the ‘specialized’ data.
Extended Data Fig. 3 Prediction of median object diameter.
a, The style vectors were used as linear regressors to predict the diameter of the objects in each image. Shown are the predictions for 68 test images (specialized and generalized data together), which were not used either for training cellpose or for training the size model. b, The refined size predictions are obtained after running cellpose on images resized according to the sizes computed in a. The median diameter of resulting objects is used as the refined size prediction for the final cellpose run. cd, Same as ab for the nucleus dataset.
Extended Data Fig. 4 Example Stardist segmentations.
Compare to Fig. 4.
Extended Data Fig. 5 Example Mask R-CNN segmentations.
Compare to Fig. 4.
Extended Data Fig. 6 Other performance metrics.
ab, Boundary prediction metrics (precision, recall, F-score42) for: a, specialized and b, generalized data. All images of masks were first resized to an average ROI diameter of 30 pixels so that a common boundary width can be used across images. cd, Aggregated Jaccard Index43 for: c, specialized (n=11 test images) and d, generalized data (n=57 test images). **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; Wilcoxon two-sided signed-rank test.
Extended Data Fig. 7 Benchmarks for dataset of nuclei.
a, Embedding of image styles for the nuclear dataset of 1139 images, with a new Cellpose model trained on this dataset. Legend: dark blue = Data Science Bowl dataset, blue = extra kaggle, cyan = ISBI 2009, green = MoNuSeg. b, Segmentations on one example test image. c, Accuracy precision scores on test data for Cellpose, Mask R-CNN, Stardist, unet3, and unet2 on n=111 test images.
Extended Data Fig. 8 Example cellpose segmentations for nuclei.
The predicted masks are shown in dotted yellow line.
Supplementary information
Supplementary Information
Supplementary Table 1.
Supplementary Video 1
xy projections through 3D volume with superimposed Cellpose3D ROI.
Supplementary Video 2
Rotating Cellpose3D ROI.
Rights and permissions
About this article
Cite this article
Stringer, C., Wang, T., Michaelos, M. et al. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100–106 (2021). https://doi.org/10.1038/s41592-020-01018-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-020-01018-x
This article is cited by
-
pyHiM: a new open-source, multi-platform software package for spatial genomics based on multiplexed DNA-FISH imaging
Genome Biology (2024)
-
Bento: a toolkit for subcellular analysis of spatial transcriptomics data
Genome Biology (2024)
-
Automatic 3D cell segmentation of fruit parenchyma tissue from X-ray micro CT images using deep learning
Plant Methods (2024)
-
Spatial insights into immunotherapy response in non-small cell lung cancer (NSCLC) by multiplexed tissue imaging
Journal of Translational Medicine (2024)
-
Slideflow: deep learning for digital histopathology with real-time whole-slide visualization
BMC Bioinformatics (2024)