Abstract
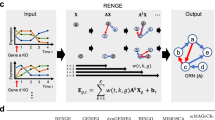
We present a systematic evaluation of state-of-the-art algorithms for inferring gene regulatory networks from single-cell transcriptional data. As the ground truth for assessing accuracy, we use synthetic networks with predictable trajectories, literature-curated Boolean models and diverse transcriptional regulatory networks. We develop a strategy to simulate single-cell transcriptional data from synthetic and Boolean networks that avoids pitfalls of previously used methods. Furthermore, we collect networks from multiple experimental single-cell RNA-seq datasets. We develop an evaluation framework called BEELINE. We find that the area under the precision-recall curve and early precision of the algorithms are moderate. The methods are better in recovering interactions in synthetic networks than Boolean models. The algorithms with the best early precision values for Boolean models also perform well on experimental datasets. Techniques that do not require pseudotime-ordered cells are generally more accurate. Based on these results, we present recommendations to end users. BEELINE will aid the development of gene regulatory network inference algorithms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The datasets simulated from the synthetic networks and curated models and the processed experimental single-cell gene expression datasets are available on Zenodo at https://doi.org/10.5281/zenodo.3378975. The gene experimental scRNA-seq datasets we downloaded from Gene Expression Omnibus had the accession numbers GSE81252 (hHEP), GSE75748 (hESC), GSE98664 (mESC), GSE48968 (mouse dendritic cell) and GSE81682 (mHSC). Source data for Figs. 2 and 4–6 are provided with the paper.
Code availability
A Python implementation of the BEELINE framework is available under the GNU General Public License v.3 at https://github.com/murali-group/BEELINE.
References
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Rozenblatt-Rosen, O., Stubbington, M. J. T., Regev, A. & Teichmann, S. A. The human cell atlas: from vision to reality. Nature 550, 451–453 (2017).
Wagner, A., Regev, A. & Yosef, N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 34, 1145–1160 (2016).
Kharchenko, P. V., Silberstein, L. & Scadden, D. T. Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742 (2014).
Buettner, F. et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 33, 155–160 (2015).
Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS One 5, e12776 (2010).
Kim, S. ppcor: An R package for a fast calculation to semi-partial correlation coefficients. Commun. Stat. Appl. Methods 22, 665–674 (2015).
Moerman, T. et al. GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics 35, 2159–2161 (2018).
Aubin-Frankowski, P.-C. & Vert, J.-P. Gene regulation inference from single-cell RNA-seq data with linear differential equations and velocity inference. Preprint at bioRxiv https://doi.org/10.1101/464479 (2018).
Deshpande, A., Chu, L.-F., Stewart, R. & Gitter, A. Network inference with Granger causality ensembles on single-cell transcriptomic data. Preprint at bioRxiv https://doi.org/10.1101/534834 (2019).
Huynh-Thu, V. A. & Sanguinetti, G. Combining tree-based and dynamical systems for the inference of gene regulatory networks. Bioinformatics 31, 1614–1622 (2015).
Specht, A. T. & Li, J. LEAP: constructing gene co-expression networks for single-cell RNA-sequencing data using pseudotime ordering. Bioinformatics 33, 764–766 (2017).
Matsumoto, H. et al. SCODE: an efficient regulatory network inference algorithm from single-cell RNA-Seq during differentiation. Bioinformatics 33, 2314–2321 (2017).
Chan, T. E., Stumpf, M. P. H. & Babtie, A. C. Gene regulatory network inference from single-cell data using multivariate information measures. Cell Syst. 5, 251–267 (2017).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Papili Gao, N., Ud-Dean, S. M. M., Gandrillon, O. & Gunawan, R. SINCERITIES: inferring gene regulatory networks from time-stamped single cell transcriptional expression profiles. Bioinformatics 34, 258–266 (2018).
Sanchez-Castillo, M., Blanco, D., Tienda-Luna, I. M., Carrion, M. C. & Huang, Y. A Bayesian framework for the inference of gene regulatory networks from time and pseudo-time series data. Bioinformatics 34, 964–970 (2018).
Woodhouse, S., Piterman, N., Wintersteiger, C. M., Göttgens, B. & Fisher, J. SCNS: a graphical tool for reconstructing executable regulatory networks from single-cell genomic data. BMC Syst. Biol. 12, 59 (2018).
Qiu, X. et al. Towards inferring causal gene regulatory networks from single cell expression measurements. Preprint at bioRxiv https://doi.org/10.1101/426981 (2018).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019).
Lim, C. Y. et al. BTR: training asynchronous Boolean models using single-cell expression data. BMC Bioinforma. 17, 355 (2016).
Chen, S. & Mar, J. C. Evaluating methods of inferring gene regulatory networks highlights their lack of performance for single cell gene expression data. BMC Bioinformatics 19, 232 (2018).
Schaffter, T., Marbach, D. & Floreano, D. GeneNetWeaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics 27, 2263–2270 (2011).
Ocone, A., Haghverdi, L., Mueller, N. S. & Theis, F. J. Reconstructing gene regulatory dynamics from high-dimensional single-cell snapshot data. Bioinformatics 31, 89–96 (2015).
Giacomantonio, C. E. & Goodhill, G. J. A Boolean model of the gene regulatory network underlying mammalian cortical area development. PLoS Comput. Biol. 6, e1000936 (2010).
Lovrics, A. et al. Boolean modelling reveals new regulatory connections between transcription factors orchestrating the development of the ventral spinal cord. PLoS One 9, e111430 (2014).
Krumsiek, J., Marr, C., Schroeder, T. & Theis, F. J. Hierarchical differentiation of myeloid progenitors is encoded in the transcription tactor network. PLoS One 6, e22649 (2011).
Ríos, O. et al. A Boolean network model of human gonadal sex determination. Theor. Biol. Med. Model. 12, 26 (2015).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Marbach, D. et al. Wisdom of crowds for robust gene network inference. Nat. Methods 9, 796–804 (2012).
Luecken, M. D. & Theis, F. J. Current best practices in single‐cell RNA‐seq analysis: a tutorial. Mol. Syst. Biol. 15, 8746 (2019).
Svensson, V., Vento-Tormo, R. & Teichmann, S. A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 13, 599–604 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Mimitou, E. P. et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019).
Faith, J. J. et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5, e8 (2007).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Yuan, Y. & Bar-Joseph, Z. Deep learning for inferring gene relationships from single-cell expression data. Preprint at bioRxiv https://doi.org/10.1101/365007 (2019).
Chen, H. et al. Single-cell transcriptional analysis to uncover regulatory circuits driving cell fate decisions in early mouse development. Bioinformatics 31, 1060–1066 (2015).
Hamey, F. K. et al. Reconstructing blood stem cell regulatory network models from single-cell molecular profiles. Proc. Natl Acad. Sci. USA 114, 5822–5829 (2017).
Marbach, D., Schaffter, T., Mattiussi, C. & Floreano, D. Generating realistic in silico gene networks for performance assessment of reverse engineering methods. J. Comput. Biol. 16, 229–239 (2009).
Marbach, D. et al. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl Acad. Sci. USA 107, 6286–6291 (2010).
Ackers, G. K., Johnson, A. D. & Shea, M. A. Quantitative model for gene regulation by λ phage repressor. Proc. Natl Acad. Sci. USA 79, 1129–1133 (1982).
Liu, Z. P., Wu, C., Miao, H. & Wu, H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database 2015, bav095 (2015).
Han, H. et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 46, D380–D386 (2018).
Garcia-Alonso, L., Holland, C. H., Ibrahim, M. M., Turei, D. & Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 29, 1363–1375 (2019).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, 607–613 (2018).
Brennecke, P. et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 10, 1093–1095 (2013).
Nestorowa, S. et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, 20–31 (2016).
Hayashi, T. et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat. Commun. 9, 619 (2018).
Shalek, A. K. et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014).
Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017).
Chu, L. F. et al. Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol. 17, 173 (2016).
Saito, T. & Rehmsmeier, M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 10, e0118432 (2015).
Saito, T. & Rehmsmeier, M. Precrec: fast and accurate precision-recall and ROC curve calculations in R. Bioinformatics 33, 145–147 (2017).
Acknowledgements
Grants from the National Science Foundation (nos. CCF-1617678 and DBI-1759858) and the National Cancer Institute (grant no. UH2CA203768) supported this work. The research is also based on work supported by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via the Army Research Office (ARO) under cooperative agreement no. W911NF-17-2-0105. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the ODNI, IARPA, ARO or the US Government. The US Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright annotation thereon. The funding bodies played no role in the design of the study, the collection, analysis and interpretation of data or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
A.P. and T.M.M. conceived and designed the analysis and selected the GRN algorithms. A.P. implemented BEELINE and led the analysis. A.P.J., A.P. and T.M.M. developed BoolODE and A.P.J. implemented it. A.P. and A.P.J. created and processed datasets. A.P., A.P.J., J.N.L. and A.B. contributed evaluation strategies. All authors analyzed the results. A.P., A.P.J. and T.M.M. wrote the paper. T.M.M. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nicole Rusk and Lin Tang were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Comparison of datasets simulated from synthetic networks by using BoolODE and GeneNetWeaver.
Each row corresponds to the synthetic network indicated by the label on the left. (a) The network itself, with red edges representing inhibition and blue edges representing activation. (b) A 2D t-SNE visualization of one BoolODE-generated dataset for 2,000 cells. The color of each point indicates the simulation time: blue for earlier, green for intermediate, and yellow for later times. (c) Each colour corresponds to a different subset of cells obtained by using k-means clustering of the BoolODE-generated dataset, with k set to the number of expected steady states. (d) A 2-D t-SNE visualization of one GeneNetWeaver output.
Supplementary Figure 2 Box plots of AUPRC values for synthetic networks.
Each row corresponds to one of the six synthetic networks. Each column corresponds to an algorithm. Red, blue, yellow, purple and green box plots correspond to AUPRC values for 10 datasets with 100, 200, 500, 2,000, and 5,000 cells, respectively. The gray dotted line indicates the AUPRC value for a random predictor, which is equal to the network’s density. In every boxplot, the box shows the 1st and 3rd quartile, and whiskers denote 1.5 times the interquartile range.
Supplementary Figure 3 Box plots of AUROC values for synthetic networks.
Each row corresponds to one of the six synthetic networks. Each column corresponds to an algorithm. Red, blue, yellow, purple and green box plots correspond to AUROC values for 10 datasets with 100, 200, 500, 2,000, and 5,000 cells, respectively. The gray dotted line indicates the AUROC value for a random predictor (0.5). In every boxplot, the box shows the 1st and 3rd quartile, and whiskers denote 1.5 times the interquartile range.
Supplementary Figure 4 Box plots of AUPRC values for curated models.
Each row corresponds to one of the four curated models. Each column corresponds to an algorithm. Red, blue and yellow box plots correspond to AUPRC values for 10 datasets with no dropouts, a dropout rate of q = 50, and a dropout rate of q = 70, respectively. The gray dotted line indicates the AUPRC value for a random predictor, i.e., the network density. In every boxplot, the box shows the 1st and 3rd quartile, and whiskers denote 1.5 times the interquartile range.
Supplementary Figure 5 Box plots of AUROC values for curated models.
Each row corresponds to one of the four curated models. Each column corresponds to an algorithm. Red, blue and yellow box plots correspond to AUROC values for 10 datasets with no dropouts, a dropout rate of q = 50, and a dropout rate of q = 70, respectively. The gray dotted line indicates the AUROC value for a random predictor (0.5). In all boxplots, the box shows the 1st and 3rd quartile, and whiskers denote 1.5 times the interquartile range.
Supplementary Figure 6 Box plots of early precision values for curated models.
Each row corresponds to one of the four curated models. Each column corresponds to an algorithm. Red, blue and yellow box plots correspond to early precision values for 10 datasets with no dropouts, a dropout rate of q = 50, and a dropout rate of q = 70, respectively. The gray dotted line indicates the early precision value for a random predictor (network density). In each boxplot, the box shows the 1st and 3rd quartile, and whiskers denote 1.5 times the interquartile range.
Supplementary Figure 7 Scalability of GRN algorithms on experimental single-cell RNA-Seq datasets.
Variation in running time and memory usage of GRN inference algorithms with respect to number of genes for three experimental single-cell RNA-Seq datasets. Each point represents the mean running time or memory across all three datasets and the shaded regions correspond to one standard deviation around the mean. Missing values indicate that the method either did not complete after one day or gave a runtime error. We did not consider SCNS since it took over a day on the 19-gene GSD Boolean model. We obtained these results on a computer with a 32-core 2.0GHz processor and 32GB of memory running Ubuntu 18.04.
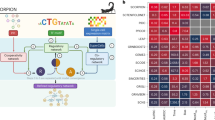
Supplementary Figure 8 Summary of EPR values for experimental single-cell RNA-Seq datasets with 500 and 1000 genes.
Summary of EPR results for experimental single-cell RNA-seq datasets. The left half of the figure (500 genes) shows results for datasets composed of the 500 most-varying genes. Each row corresponds to one scRNA-seq dataset. The first three columns report network statistics. The next six columns report EPR values. The right half (1000 genes) shows results for the 1000 most-varying genes. In both sections, algorithms are sorted by median EPR across the datasets (rows) for the 500 gene set. For each dataset, the color in each cell is proportional to the corresponding value scaled between 0 and 1 (ignoring values that are less than that of a random predictor, which are shown as black squares). We display the highest and lowest values for each dataset inside the corresponding cells. Abbreviations: GENI: GENIE3, GRNB: GRNBoost2, PCOR: PPCOR, SINC: SINCERITIES.
Supplementary Figure 9 Summary of AUPRC ratio values for experimental single-cell RNA-Seq datasets with TFs + 500 and TFs + 1000 genes.
Summary of AUPRC ratio results for experimental single-cell RNA-seq datasets. The left half of the figure (TFs+500 genes) shows results for datasets composed of all significantly-varying TFs and the 500 most-varying genes. Each row corresponds to one scRNA-seq dataset. The first three columns report network statistics. The next six columns report AUPRC ratios. The right half (TFs+1000 genes) shows results for all significantly-varying TFs and the 1000 most-varying genes. In both sections, algorithms are sorted by median AUPRC ratio across the datasets (rows) for the TFs+500 gene set. For each dataset, the color in each cell is proportional to the corresponding value scaled between 0 and 1 (ignoring values that are less than that of a random predictor, which are shown as black squares). We display the highest and lowest values for each dataset inside the corresponding cells. Abbreviations: GENI: GENIE3, GRNB: GRNBoost2, PCOR: PPCOR, SINC: SINCERITIES.
Supplementary Figure 10 Summary of AUPRC ratio values for experimental single-cell RNA-Seq datasets with 500 and 1000 genes.
Summary of AUPRC ratio values for experimental single-cell RNA-seq datasets. The left half of the figure (500 genes) shows results for datasets composed of the 500 most-varying genes. Each row corresponds to one scRNA-seq dataset. The first three columns report network statistics. The next six columns report AUPRC ratios. The right half (1000 genes) shows results for the 1000 most-varying genes. In both sections, algorithms are sorted by median AUPRC ratios across the datasets (rows) for the 500 gene set. For each dataset, the color in each cell is proportional to the corresponding value scaled between 0 and 1 (ignoring values that are less than that of a random predictor, which are shown as black squares). We display the highest and lowest values for each dataset inside the corresponding cells. Abbreviations: GENI: GENIE3, GRNB: GRNBoost2, PCOR: PPCOR, SINC: SINCERITIES.
Supplementary Figure 11 Effect of pseudotime shuffling on AUPRC values for synthetic networks.
(a) Comparison of AUPRC values for networks inferred with original pseudotimes (blue) and shuffled within three window sizes 15% (light blue), 30% (beige) or 45% (orange). Each row corresponds to one of the three synthetic networks with single trajectories (Linear, Cycle, and Linear Long). Each boxplot represents 10 AUPRC values (10 datasets each containing 2,000 cells). The gray dotted line indicates the AUPRC value for a random predictor (i.e., the network density). In each boxplot, the box shows the 1st and 3rd quartile, and whiskers denote 1.5 times the interquartile range. (b) Median Pearson correlation between original and shuffled pseudotimes. For each network and window-size the median is over 10 datasets.
Supplementary information
Supplementary Information
Supplementary Figs. 1–11 and notes.
Supplementary Table 1
Summary of synthetic networks
Supplementary Table 2
Summary of published Boolean models.
Supplementary Table 3
Statistics on experimental single-cell RNA-seq datasets.
Supplementary Table 4
Statistics on networks used to evaluate experimental single-cell RNA-seq datasets
Supplementary Table 5
Truth table and parameter values for the BoolODE example.
Supplementary Table 6
Kinetic parameters used in BoolODE.
Supplementary Table 7
Parameters used to generate synthetic datasets.
Supplementary Table 8
Median Pearson correlation between simulation time and pseudotime computed using Slingshot for different dropout rates.
Supplementary Table 9
Software version details for the 12 GRN inference algorithms in BEELINE.
Source data
Source Data Fig. 2
Synthetic networks.
Source Data Fig. 4
Curated models.
Source Data Fig. 5
Experimental.
Source Data Fig. 6
Summary.
Rights and permissions
About this article
Cite this article
Pratapa, A., Jalihal, A.P., Law, J.N. et al. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat Methods 17, 147–154 (2020). https://doi.org/10.1038/s41592-019-0690-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0690-6
This article is cited by
-
PMF-GRN: a variational inference approach to single-cell gene regulatory network inference using probabilistic matrix factorization
Genome Biology (2024)
-
scDisInFact: disentangled learning for integration and prediction of multi-batch multi-condition single-cell RNA-sequencing data
Nature Communications (2024)
-
scPerturb: harmonized single-cell perturbation data
Nature Methods (2024)
-
Challenges and best practices in omics benchmarking
Nature Reviews Genetics (2024)
-
Enabling comparative gene regulatory network analysis on single-cell data with SCORPION
Nature Computational Science (2024)