Abstract
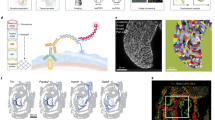
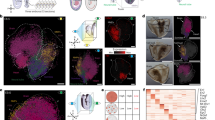
The Drosophila wing disc has been a fundamental model system for the discovery of key signaling pathways and for our understanding of developmental processes. However, a complete map of gene expression in this tissue is lacking. To obtain a gene expression atlas in the wing disc, we employed single cell RNA sequencing (scRNA-seq) and developed a method for analyzing scRNA-seq data based on gene expression correlations rather than cell mapping. This enables us to compute expression maps for all detected genes in the wing disc and to discover 824 genes with spatially restricted expression patterns. This approach identifies clusters of genes with similar expression patterns and functional relevance. As proof of concept, we characterize the previously unstudied gene CG5151 and show that it regulates Wnt signaling. Our method will enable the leveraging of scRNA-seq data for generating expression atlases of undifferentiated tissues during development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data generated or analyzed during this study are included in this published article (and its Supplementary Information files). Raw sequencing data have been deposited to NCBI GEO with accession number GSE127832.
Code availability
All custom code, together with sample data, are freely available on the Boutros laboratory repository of Github (https://github.com/boutroslab/Supplemental-Material/tree/master/Bageritz_2019). A markdown document describing how to identify SRGs using R is provided. The software package is composed of four pieces of software: (1) 1_cross_correlation_all_genes calculates the cross-correlation of all genes against all genes provided as input in an expression matrix. (2) 2_identify_best_mapping_genes recursively identifies the gene with the highest correlation score in the cross-correlation matrix generated by software no. 1, and pulls it out as a mapping gene. See Supplementary Fig. 12b for a schematic diagram. (3) 3_cross_correlation_to_mapping_genes calculates the cross-correlation between all genes and the SRGs, taking as input an expression matrix. (4) 4_calculate_expression_maps calculates the expression maps following the algorithm described above in the section Generation of computed wing disc maps.
References
Buchmann, A., Alber, M. & Zartman, J. J. Sizing it up: the mechanical feedback hypothesis of organ growth regulation. Semin. Cell Dev. Biol. 35C, 73–81 (2014).
Restrepo, S., Zartman, J. J. & Basler, K. Coordination of patterning and growth by the morphogen DPP. Curr. Biol. 24, R245–R255 (2014).
Worley, M. I., Setiawan, L. & Hariharan, I. K. Regeneration and transdetermination in Drosophila imaginal discs. Annu. Rev. Genet. 46, 289–310 (2012).
Garcia-Bellido, A. The cellular and genetic bases of organ size and shape in Drosophila. Int. J. Dev. Biol. 53, 1291–1303 (2009).
Kornberg, T. B. & Guha, A. Understanding morphogen gradients: a problem of dispersion and containment. Curr. Opin. Genet. Dev. 17, 264–271 (2007).
LeGoff, L. & Lecuit, T. Mechanical forces and growth in animal tissues. Cold Spring Harb. Perspect. Biol. 8, a019232 (2015).
Johnston, L. A. Socializing with MYC: cell competition in development and as a model for premalignant cancer. Cold Spring Harb. Perspect. Med. 4, a014274 (2014).
Calleja, M., Moreno, E., Pelaz, S. & Morata, G. Visualization of gene expression in living adult Drosophila. Science 274, 252–255 (1996).
Ibrahim, D. M., Biehs, B., Kornberg, T. B. & Klebes, A. Microarray comparison of anterior and posterior Drosophila wing imaginal disc cells identifies novel wing genes. G3 3, 1353–1362 (2013).
Reeves, N. & Posakony, J. W. Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev. Cell 8, 413–425 (2005).
Svensson, V., Teichmann, S. A. & Stegle, O. SpatialDE: identification of spatially variable genes. Nat. Meth. 15, 343–346 (2018).
Edsgard, D., Johnsson, P. & Sandberg, R. Identification of spatial expression trends in single-cell gene expression data. Nat. Meth. 15, 339–342 (2018).
Davie, K. et al. A single-cell transcriptome atlas of the aging drosophila brain. Cell 174, 982–998 (2018).
Han, X. et al. Mapping the mouse cell atlas by microwell-seq. Cell 173, 1307 (2018).
Croset, V., Treiber, C. D. & Waddell, S. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. eLife 7, e34550 (2018).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Karaiskos, N. et al. The Drosophila embryo at single-cell transcriptome resolution. Science 358, 194–199 (2017).
Achim, K. et al. High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat. Biotechnol. 33, 503–509 (2015).
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017).
Zhu, Q., Shah, S., Dries, R., Cai, L. & Yuan, G. C. Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data. Nat. Biotech. 36, 1183–1190 (2018).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Andrews, T. S. & Hemberg, M. M3Drop: Dropout-based feature selection for scRNASeq. Bioinformatics https://doi.org/10.1093/bioinformatics/bty1044 (2018).
Morikawa, R. K., Kanamori, T., Yasunaga, K. & Emoto, K. Different levels of the tripartite motif protein, anomalies in sensory axon patterning (ASAP), regulate distinct axonal projections of Drosophila sensory neurons. Proc. Natl Acad. Sci. USA 108, 19389–19394 (2011).
Lammel, U. et al. The Drosophila FHOD1-like formin Knittrig acts through Rok to promote stress fiber formation and directed macrophage migration during the cellular immune response. Development 141, 1366–1380 (2014).
Sieglitz, F. et al. Antagonistic feedback loops involving Rau and Sprouty in the Drosophila eye control neuronal and glial differentiation. Sci. Signal. 6, ra96 (2013).
Glasscock, E. & Tanouye, M. A. Drosophila couch potato mutants exhibit complex neurological abnormalities including epilepsy phenotypes. Genetics 169, 2137–2149 (2005).
Bellen, H. J., Kooyer, S., D’Evelyn, D. & Pearlman, J. The Drosophila couch potato protein is expressed in nuclei of peripheral neuronal precursors and shows homology to RNA-binding proteins. Genes Dev. 6, 2125–2136 (1992).
Clohisey, S. M., Dzhindzhev, N. S. & Ohkura, H. Kank Is an EB1 interacting protein that localises to muscle-tendon attachment sites in Drosophila. PloS ONE 9, e106112 (2014).
Liu, Z. et al. Low density lipoprotein receptor class A domain containing 4 (LDLRAD4) promotes tumorigenesis of hepatic cancer cells. Exp. Cell Res. 360, 189–198 (2017).
Su, G., Morris, J. H., Demchak, B. & Bader, G. D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinforma. 47, 11–24 (2014). 8 13.
McCarthy, D. J., Campbell, K. R., Lun, A. T. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Acknowledgements
We thank the High Throughput Sequencing group of the DKFZ Genomics and Proteomics Core Facility for providing excellent next-generation sequencing services. We thank J.-P. Mallm and the DKFZ Single-Cell Open Laboratory (scOpenLab) for assistance with the 10X Genomics experiment. We thank E. Rempel for implementing the DropSeq computational cookbook on our Galaxy Server. J.B. was supported by a research stipend from the Fritz Thyssen Foundation. P.W. was funded by a fellowship from CellNetworks—Cluster of Excellence (EXC81). Research in the laboratories of M.B. and A.A.T. is supported by ERC Grants of the European Commission.
Author information
Authors and Affiliations
Contributions
J.B., P.W., S.L. and A.A.T. performed experiments. J.B., P.W., E.V., M.B. and A.A.T. analyzed data. J.B., P.W., M.B. and A.A.T. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Nina Vogt and Tal Nawy were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs 1–15 and Supplementary Table 1.
Supplementary Table 2
Spatially Regulated Genes (SRGs) in the wing disc. List of 824 SRGs identified by scRNA-seq of wing disc cells.
Supplementary Table 3
‘Benchmark’ genes with spatially restricted expression domains in the wing disc. 68 genes known from literature to have spatially restricted expression patterns in the wing disc.
Supplementary Table 4
Mapping genes. List of 58 mapping genes used for generating computed expression maps of the Drosophila wing disc.
Supplementary Table 5
Genes correlating to Senseless. n=948 wing disc cells. Correlation was calculated using Pearson’s correlation coefficient with one outlier removed. See Methods for details.
Supplementary Table 6
Genes correlating to Wingless. n=948 wing disc cells. Correlation was calculated using Pearson’s correlation coefficient with one outlier removed. See Methods for details.
Supplementary Table 7
Genes correlating to Dpp. n=948 wing disc cells. Correlation was calculated using Pearson’s correlation coefficient with one outlier removed. See Methods for details.
Supplementary Table 8
Best mapping genes for the wing disc identified de novo from scRNA-seq data. n=948 wing disc cells. Correlation was calculated using Pearson’s correlation coefficient with one outlier removed. See Methods for details.
Supplementary Table 9
Best mapping genes for Drosophila embryo (stage 5) identified de novo from scRNA-seq data.
Supplementary Table 10
Oligo sequences. Sequences of oligos used to generate probes for in situs.
Supplementary Data Set 1
High-resolution version of the dendogram presented in Fig. 3a. This version allows the names of all the genes to be read by zooming in.
Supplementary Data Set 2
Computed expression maps for all SRGs. This compressed file contains a folder with computed expression maps for all SRGs in the wing disc as TIFF image files.
Supplementary Software
Compressed ZIP file containing executable software (as MacOS binaries), source code (in C) and sample data.
Rights and permissions
About this article
Cite this article
Bageritz, J., Willnow, P., Valentini, E. et al. Gene expression atlas of a developing tissue by single cell expression correlation analysis. Nat Methods 16, 750–756 (2019). https://doi.org/10.1038/s41592-019-0492-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0492-x
This article is cited by
-
Explainable multiview framework for dissecting spatial relationships from highly multiplexed data
Genome Biology (2022)
-
A novel graph-based k-partitioning approach improves the detection of gene-gene correlations by single-cell RNA sequencing
BMC Genomics (2022)
-
Statistical and machine learning methods for spatially resolved transcriptomics data analysis
Genome Biology (2022)
-
Genome-wide analysis of MYB transcription factors of Vaccinium corymbosum and their positive responses to drought stress
BMC Genomics (2021)
-
Model-based prediction of spatial gene expression via generative linear mapping
Nature Communications (2021)