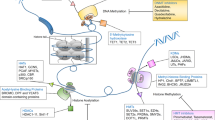
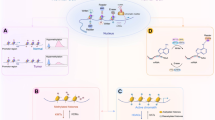
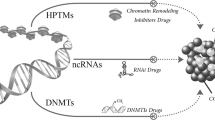
Abstract
Epigenetic dysregulation is a common feature of most cancers, often occurring directly through alteration of epigenetic machinery. Over the last several years, a new generation of drugs directed at epigenetic modulators have entered clinical development, and results from these trials are now being disclosed. Unlike first-generation epigenetic therapies, these new agents are selective, and many are targeted to proteins which are mutated or translocated in cancer. This review will provide a summary of the epigenetic modulatory agents currently in clinical development and discuss the opportunities and challenges in their development. As these drugs advance in the clinic, drug discovery has continued with a focus on both novel and existing epigenetic targets. We will provide an overview of these efforts and the strategies being employed.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Allis, C. D. & Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016).
Dubuc, A. M. et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 125, 373–384 (2013).
Fontebasso, A. M. et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 125, 659–669 (2013).
Gui, Y. et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 43, 875–878 (2011).
Jones, D. T. et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 488, 100–105 (2012).
Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 (2010).
Kim, J. H. et al. Deep sequencing reveals distinct patterns of DNA methylation in prostate cancer. Genome Res. 21, 1028–1041 (2011).
Le Gallo, M. et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 44, 1310–1315 (2012).
Lee, W. et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 46, 1227–1232 (2014).
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
Zang, Z. J. et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat. Genet. 44, 570–574 (2012).
Pfister, S. X. & Ashworth, A. Marked for death: targeting epigenetic changes in cancer. Nat. Rev. Drug Discov. 16, 241–263 (2017).
Jones, P. A. & Taylor, S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85–93 (1980).
Constantinides, P. G., Taylor, S. M. & Jones, P. A. Phenotypic conversion of cultured mouse embryo cells by aza pyrimidine nucleosides. Dev. Biol. 66, 57–71 (1978).
Sorm, F. & Veselý, J. Effect of 5-aza-2′-deoxycytidine against leukemic and hemopoietic tissues in AKR mice. Neoplasma 15, 339–343 (1968).
Yoshida, M., Nomura, S. & Beppu, T. Effects of trichostatins on differentiation of murine erythroleukemia cells. Cancer Res. 47, 3688–3691 (1987).
Vidali, G., Boffa, L. C., Bradbury, E. M. & Allfrey, V. G. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc. Natl. Acad. Sci. USA 75, 2239–2243 (1978).
Stahl, M., Gore, S. D., Vey, N. & Prebet, T. Lost in translation? Ten years of development of histone deacetylase inhibitors in acute myeloid leukemia and myelodysplastic syndromes. Expert. Opin. Investig. Drugs 25, 307–317 (2016).
Chitambar, C. R. et al. Evaluation of continuous infusion low-dose 5-azacytidine in the treatment of myelodysplastic syndromes. Am. J. Hematol. 37, 100–104 (1991).
Issa, J. P. et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 103, 1635–1640 (2004).
Issa, J. P. & Kantarjian, H. M. Targeting DNA methylation. Clin. Cancer Res. 15, 3938–3946 (2009).
Whittaker, S. J. et al. Final results from a multicenter, international, pivotal study of romidepsin in refractory cutaneous T-cell lymphoma. J. Clin. Oncol. 28, 4485–4491 (2010).
Duvic, M. et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL). Blood 109, 31–39 (2007).
Prebet, T. et al. Prolonged administration of azacitidine with or without entinostat for myelodysplastic syndrome and acute myeloid leukemia with myelodysplasia-related changes: results of the US Leukemia Intergroup trial E1905. J. Clin. Oncol. 32, 1242–1248 (2014).
Silverman, L. R. et al. A phase II trial of epigenetic modulators vorinostat in combination with azacitidine (azaC) in patients with the myelodysplastic syndrome (MDS): initial results of Study 6898 of the New York Cancer Consortium. Blood 122, 386 (2013).
Tan, P. et al. Dual epigenetic targeting with panobinostat and azacitidine in acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood Cancer J. 4, e170 (2014).
Linnekamp, J. F., Butter, R., Spijker, R., Medema, J. P. & van Laarhoven, H. W. M. Clinical and biological effects of demethylating agents on solid tumours - a systematic review. Cancer Treat. Rev. 54, 10–23 (2017).
Suraweera, A., O’Byrne, K. J. & Richard, D. J. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front. Oncol. 8, 92 (2018).
Wiegand, K. C. et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543 (2010).
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 41, 521–523 (2009).
Nikoloski, G. et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 42, 665–667 (2010).
Ernst, T. et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 42, 722–726 (2010).
Chiba, S. Dysregulation of TET2 in hematologic malignancies. Int. J. Hematol. 105, 17–22 (2017).
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 42, 181–185 (2010).
Russler-Germain, D. A. et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell 25, 442–454 (2014).
Parikh, S. A. et al. NUT midline carcinoma: an aggressive intrathoracic neoplasm. J. Thorac. Oncol. 8, 1335–1338 (2013).
Kuo, A. J. et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 44, 609–620 (2011).
Agrawal, K., Das, V., Vyas, P. & Hajdúch, M. Nucleosidic DNA demethylating epigenetic drugs - a comprehensive review from discovery to clinic. Pharmacol. Ther. 188, 45–79 (2018).
Fenaux, P. et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J. Clin. Oncol. 28, 562–569 (2010).
Fenaux, P. et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 10, 223–232 (2009).
Lübbert, M. et al. Decitabine improves progression-free survival in older high-risk MDS patients with multiple autosomal monosomies: results of a subgroup analysis of the randomized phase III study 06011 of the EORTC Leukemia Cooperative Group and German MDS Study Group. Ann. Hematol. 95, 191–199 (2016).
Stewart, D. J. et al. Decitabine effect on tumor global DNA methylation and other parameters in a phase I trial in refractory solid tumors and lymphomas. Clin. Cancer Res. 15, 3881–3888 (2009).
Zhang, W. et al. DNA hypomethylation-mediated activation of cancer/testis antigen 45 (CT45) genes is associated with disease progression and reduced survival in epithelial ovarian cancer. Epigenetics 10, 736–748 (2015).
Serrano, A. et al. Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2′-deoxycytidine treatment. Int. J. Cancer. 94, 243–251 (2001).
Cruickshank, B. et al. Dying to be noticed: epigenetic regulation of immunogenic cell death for cancer immunotherapy. Front. Immunol. 9, 654 (2018).
Yang, H. et al. Expression of PD-L1, PD-L2, PD-1 and CTLA4 in myelodysplastic syndromes is enhanced by treatment with hypomethylating agents. Leukemia 28, 1280–1288 (2014).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Roulois, D. et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
Issa, J. J. et al. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol. 16, 1099–1110 (2015).
Yoo, C. B. et al. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 67, 6400–6408 (2007).
Kantarjian, H. M. et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol. 18, 1317–1326 (2017).
Pappalardi, M.B., et al. Abstr. 2994: Discovery selective, noncovalent small molecule inhibitors DNMT1 an alternative traditional DNA hypomethylating Agent. Proc. AACR Annual Meeting 2018 (2018).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Bödör, C. et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood 122, 3165–3168 (2013).
Lohr, J. G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA 109, 3879–3884 (2012).
Kleer, C. G. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA 100, 11606–11611 (2003).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Yap, D. B. et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 117, 2451–2459 (2011).
Sneeringer, C. J. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA 107, 20980–20985 (2010).
Wigle, T. J. et al. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS Lett. 585, 3011–3014 (2011).
Knutson, S. K. et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13, 842–854 (2014).
Vaswani, R. G. et al. Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J. Med. Chem. 59, 9928–9941 (2016).
Kung, P. P. et al. Optimization of orally bioavailable enhancer of zeste homolog 2 (EZH2) inhibitors using ligand and property-based design strategies: identification of development candidate (R)-5,8-dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497). J. Med. Chem. 61, 650–665 (2018).
Morschhauser, F. et al. Interim report from a phase 2 multicenter study of tazemetostat, an EZH2 inhibitor, in patients with relapsed or refractory b‐cell non‐Hodgkin lymphomas. Hematol. Oncol. 35, 24–25 (2017).
Knutson, S. K. et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl Acad. Sci. USA 110, 7922–7927 (2013).
Zauderer, M. G. et al. Phase 2, multicenter study of the EZH2 inhibitor tazemetostat as monotherapy in adults with relapsed or refractory (R/R) malignant mesothelioma (MM) with BAP1 inactivation. J. Clin. Oncol. 36, 8515–8515 (2018).
Gounder, M. et al. A phase 2, multicenter study of the EZH2 inhibitor tazemetostat in adults: (epithelioid sarcoma cohort) (NCT02601950). Ann. Oncol. 29(suppl_8), viii576–viii595 (2018).
Honma, D. et al. Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 108, 2069–2078 (2017).
Maruyama, D. et al. First-in-human study of the EZH1/2 dual inhibitor DS-3201b in patients with relapsed or refractory non-Hodgkin lymphomas — preliminary results. Blood 130(Suppl 1), 1 (2017).
Kim, K. H. et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 21, 1491–1496 (2015).
Daigle, S. R. et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 122, 1017–1025 (2013).
Kühn, M. W. et al. Targeting chromatin regulators inhibits leukemogenic gene expression in NPM1 mutant leukemia. Cancer Discov. 6, 1166–1181 (2016).
Falini, B. et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 352, 254–266 (2005).
Eytan, M. et al. A phase 1 study of the DOT1L inhibitor, pinometostat (EPZ–5676), in adults with relapsed or refractory leukemia: safety, clinical activity, exposure and target inhibition. Blood 126, 2547 (2015).
Neerav Shukla, C. W. et al. Final report of phase 1 study of the DOT1L inhibitor, pinometostat (EPZ-5676), in children with relapsed or refractory MLL-r acute leukemia. Blood 128, 2780 (2016).
Stein, E. M. et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131, 2661–2669 (2018).
Waters, N. J. Preclinical pharmacokinetics and pharmacodynamics of pinometostat (epz-5676), a first-in-class, small molecule S-adenosyl methionine competitive inhibitor of DOT1L. Eur. J. Drug Metab. Pharmacokinet. 42, 891–901 (2017).
Poulard, C., Corbo, L. & Le Romancer, M. Protein arginine methylation/demethylation and cancer. Oncotarget 7, 67532–67550 (2016).
Gerhart, S. V. et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci. Rep. 8, 9711 (2018).
Drew Rasco, A. T. et al. Abstract CT038: A phase I, open-label, dose-escalation study to investigate the safety, pharmacokinetics, pharmacodynamics, and clinical activity of GSK3326595 in subjects with solid tumors and non-Hodgkin’s lymphoma. Cancer Res. 77, Supplement (2017).
Brown, P. J. & Müller, S. Open access chemical probes for epigenetic targets. Future Med. Chem. 7, 1901–1917 (2015).
Yuan, Y. et al. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem. Biol. 7, 1152–1157 (2012).
Vedadi, M. et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 7, 566–574 (2011).
Chang, Y. et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat. Struct. Mol. Biol. 16, 312–317 (2009).
Pappano, W. N. et al. The histone methyltransferase inhibitor A-366 uncovers a role for G9a/GLP in the epigenetics of leukemia. PLoS One 10, e0131716 (2015).
Eigl, B. J. et al. A phase II study of the HDAC inhibitor SB939 in patients with castration resistant prostate cancer: NCIC clinical trials group study IND195. Invest. New Drugs 33, 969–976 (2015).
Evens, A. M. et al. A phase I/II multicenter, open-label study of the oral histone deacetylase inhibitor abexinostat in relapsed/refractory lymphoma. Clin. Cancer Res. 22, 1059–1066 (2016).
Guzman, M. L. et al. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Mol. Cancer Ther. 13, 1979–1990 (2014).
Qian, C. et al. Cancer network disruption by a single molecule inhibitor targeting both histone deacetylase activity and phosphatidylinositol 3-kinase signaling. Clin. Cancer Res. 18, 4104–4113 (2012).
Knipstein, J. & Gore, L. Entinostat for treatment of solid tumors and hematologic malignancies. Expert Opin. Investig. Drugs 20, 1455–1467 (2011).
Galli, M. et al. A phase II multiple dose clinical trial of histone deacetylase inhibitor ITF2357 in patients with relapsed or progressive multiple myeloma. Ann. Hematol. 89, 185–190 (2010).
Furlan, A. et al. Pharmacokinetics, safety and inducible cytokine responses during a phase 1 trial of the oral histone deacetylase inhibitor ITF2357 (givinostat). Mol. Med. 17, 353–362 (2011).
Younes, A. et al. Mocetinostat for relapsed classical Hodgkin’s lymphoma: an open-label, single-arm, phase 2 trial. Lancet Oncol. 12, 1222–1228 (2011).
Brunetto, A. T. et al. First-in-human, pharmacokinetic and pharmacodynamic phase I study of Resminostat, an oral histone deacetylase inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 19, 5494–5504 (2013).
Santo, L. et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood 119, 2579–2589 (2012).
Leung, D. et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature 518, 350–354 (2015).
Shen, L. et al. Class I histone deacetylase inhibitor entinostat suppresses regulatory T cells and enhances immunotherapies in renal and prostate cancer models. PLoS One 7, e30815 (2012).
Woods, D. M. et al. HDAC inhibition upregulates PD-1 ligands in melanoma and augments immunotherapy with PD-1 blockade. Cancer Immunol. Res. 3, 1375–1385 (2015).
Pili, R. et al. Immunomodulation by entinostat in renal cell carcinoma patients receiving high-dose interleukin 2: a multicenter, single-arm, phase I/II trial (NCI-CTEP#7870). Clin. Cancer Res. 23, 7199–7208 (2017).
Sharma, S. V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010).
Heijnen, W. T., De Fruyt, J., Wierdsma, A. I., Sienaert, P. & Birkenhäger, T. K. Efficacy of tranylcypromine in bipolar depression: a systematic review. J. Clin. Psychopharmacol. 35, 700–705 (2015).
Baker, G. B., Coutts, R. T., McKenna, K. F. & Sherry-McKenna, R. L. Insights into the mechanisms of action of the MAO inhibitors phenelzine and tranylcypromine: a review. J. Psychiatry Neurosci. 17, 206–214 (1992).
Harris, W. J. et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21, 473–487 (2012).
Mohammad, H. P. et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28, 57–69 (2015).
Sankar, S. et al. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin. Cancer Res. 20, 4584–4597 (2014).
Sorna, V. et al. High-throughput virtual screening identifies novel N’-(1-phenylethylidene)-benzohydrazides as potent, specific, and reversible LSD1 inhibitors. J. Med. Chem. 56, 9496–9508 (2013).
Mould, D. P., McGonagle, A. E., Wiseman, D. H., Williams, E. L. & Jordan, A. M. Reversible inhibitors of LSD1 as therapeutic agents in acute myeloid leukemia: clinical significance and progress to date. Med. Res. Rev. 35, 586–618 (2015).
Tim Somervaille, O. S. et al. Safety, phamacokinetics (PK), pharmacodynamics (PD) and emia of Ory-1001, a first-in-class inhibitor of lysine-specific histone demethylase 1A (LSD1/KDM1A): initial results from a first-in-human phase 1 study. Blood 128, 4060 (2016).
Saleque, S., Kim, J., Rooke, H. M. & Orkin, S. H. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell 27, 562–572 (2007).
Mohammad, H. P. & Kruger, R. G. Antitumor activity of LSD1 inhibitors in lung cancer. Mol. Cell. Oncol. 3, e1117700 (2016).
McGrath, J. P. et al. Pharmacological inhibition of the histone lysine demethylase KDM1A suppresses the growth of multiple acute myeloid leukemia subtypes. Cancer Res. 76, 1975–1988 (2016).
Schenk, T. et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat. Med. 18, 605–611 (2012).
Mcallister, T. E. et al. Recent progress in histone demethylase inhibitors. J. Med. Chem. 59, 1308–1329 (2016).
Jambhekar, A., Anastas, J. N. & Shi, Y. Histone lysine demethylase inhibitors. Cold Spring Harb. Perspect. Med. 7, a026484 (2017).
Duan, L. et al. KDM4/JMJD2 histone demethylase inhibitors block prostate tumor growth by suppressing the expression of AR and BMYB-regulated genes. Chem. Biol. 22, 1185–1196 (2015).
Gehling, V. S. et al. Identification of potent, selective KDM5 inhibitors. Bioorg. Med. Chem. Lett. 26, 4350–4354 (2016).
Hatch, S. B. et al. Assessing histone demethylase inhibitors in cells: lessons learned. Epigenetics Chromatin 10, 9 (2017).
Heinemann, B. et al. Inhibition of demethylases by GSK-J1/J4. Nature 514, E1–E2 (2014).
Kruidenier, L. et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408 (2012).
Vinogradova, M. et al. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12, 531–538 (2016).
French, C. A. et al. Midline carcinoma of children and young adults with NUT rearrangement. J. Clin. Oncol. 22, 4135–4139 (2004).
Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011).
Stuhlmiller, T. J. et al. Inhibition of lapatinib-induced kinome reprogramming in ERBB2-positive breast cancer by targeting BET family bromodomains. Cell Rep. 11, 390–404 (2015).
Alekseyenko, A. A. et al. The oncogenic BRD4-NUT chromatin regulator drives aberrant transcription within large topological domains. Genes Dev. 29, 1507–1523 (2015).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Stathis, A. et al. Clinical response of carcinomas harboring the BRD4-NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 6, 492–500 (2016).
O’Dwyer, P. J. et al. Abstract CT014: GSK525762, a selective bromodomain (BRD) and extra terminal protein (BET) inhibitor: results from part 1 of a phase I/II open-label single-agent study in patients with NUT midline carcinoma (NMC) and other cancers. Cancer Res. 76, Supplement (2016).
Berthon, C. et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 3, e186–e195 (2016).
Mark Dawson, E. M. S. et al. A phase I study of GSK525762, a selective bromodomain (BRD) and extra terminal protein (BET) inhibitor: results from part 1 of phase I/II open label single agent study in patients with acute myeloid leukemia (AML). Blood 130, 1377 (2017).
Gautam Borthakur, J. E. W. et al. First-in-human study of ABBV-075 (mivebresib), a pan-inhibitor of bromodomain and extra terminal (BET) proteins, in patients (pts) with relapsed/refractory (RR) acute myeloid leukemia (AML): Preliminary data. J. Clin. Oncol. 36, suppl. 7019–7019 (2018).
Blum, K. A. et al. A phase I study of CPI-0610, a bromodomain and extra terminal protein (BET) inhibitor in patients with relapsed or refractory lymphoma. Ann. Oncol. 29, iii7–iii9 (2018).
Amorim, S. et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 3, e196–e204 (2016).
Amita Patnaik, R. D. C. et al. Phase ib/2a study of PLX51107, a small molecule BET inhibitor, in subjects with advanced hematological malignancies and solid tumors. J. Clin. Oncol. 36, 2550 (2018).
Sarina Anne Piha-Paul, J. C. S. et al. Results of the first-in-human study of ABBV-075 (mivebresib), a pan-inhibitor of bromodomain (BD) and extra terminal (BET) proteins, in patients (pts) with relapsed/refractory (R/R) solid tumors. J. Clin. Oncol. 36, 2510 (2018).
Sapna Pradyuman Patel, J. E. W. et al. Uveal melanoma patients (pts) treated with abbv-075 (mivebresib), a pan-inhibitor of bromodomain and extraterminal (BET) proteins: Results from a phase 1 study. J. Clin. Oncol. 36, e14585 (2018).
Andres Forero-Torres, S. R. et al. Preliminary results from an ongoing phase 1/2 Study of INCB057643, a bromodomain and extraterminal (BET) Protein Inhibitor, in patients (pts) with advanced malignancies. Blood 130, 4048 (2017).
Warren, K. Abstract 4960: A first-in-class highly BDII-selective BET bromodomain inhibitor. Cancer Res. 78, Suppl. (2018).
Tanaka, M. et al. Design and characterization of bivalent BET inhibitors. Nat. Chem. Biol. 12, 1089–1096 (2016).
Rhyasen, G. W. et al. AZD5153: a novel bivalent BET bromodomain inhibitor highly active against hematologic malignancies. Mol. Cancer Ther. 15, 2563–2574 (2016).
Raina, K. et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl Acad. Sci. USA 113, 7124–7129 (2016).
Winter, G. E. et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol. Cell 67, 5–18 e19 (2017).
Qin, C. et al. Discovery of QCA570 as an exceptionally potent and efficacious proteolysis targeting chimera (PROTAC) degrader of the bromodomain and extra-terminal (BET) proteins capable of inducing complete and durable tumor regression. J. Med. Chem. 61, 6685–6704 (2018).
Iyer, N. G., Ozdag, H. & Caldas, C. p300/CBP and cancer. Oncogene 23, 4225–4231 (2004).
Kadoch, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013).
Scotto, L. et al. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol. Cancer 7, 58 (2008).
Kandoth, C. et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013).
Borah, J. C. et al. A small molecule binding to the coactivator CREB-binding protein blocks apoptosis in cardiomyocytes. Chem. Biol. 18, 531–541 (2011).
Chen, P. et al. Discovery and characterization of GSK2801, a selective chemical probe for the bromodomains BAZ2A and BAZ2B. J. Med. Chem. 59, 1410–1424 (2016).
Drouin, L. et al. Structure enabled design of BAZ2-ICR, a chemical probe targeting the bromodomains of BAZ2A and BAZ2B. J. Med. Chem. 58, 2553–2559 (2015).
Bamborough, P. et al. GSK6853, a chemical probe for inhibition of the BRPF1 bromodomain. ACS Med. Chem. Lett. 7, 552–557 (2016).
Demont, E. H. et al. 1,3-Dimethyl benzimidazolones are potent, selective inhibitors of the BRPF1 bromodomain. ACS Med. Chem. Lett. 5, 1190–1195 (2014).
Palmer, W. S. et al. Structure-guided design of IACS-9571, a selective high-affinity dual TRIM24-BRPF1 bromodomain inhibitor. J. Med. Chem. 59, 1440–1454 (2016).
Theodoulou, N. H. et al. Discovery of I-BRD9, a selective cell active chemical probe for bromodomain containing protein 9 inhibition. J. Med. Chem. 59, 1425–1439 (2016).
Clark, P. G. et al. LP99: discovery and synthesis of the first selective BRD7/9 bromodomain inhibitor. Angew. Chem. Int. Edn Engl. 54, 6217–6221 (2015).
Picaud, S. et al. 9H-purine scaffold reveals induced-fit pocket plasticity of the BRD9 bromodomain. J. Med. Chem. 58, 2718–2736 (2015).
Fedorov, O. et al. Selective targeting of the BRG/PB1 bromodomains impairs embryonic and trophoblast stem cell maintenance. Sci. Adv. 1, e1500723 (2015).
Moustakim, M. et al. Discovery of a PCAF bromodomain chemical probe. Angew. Chem. Int. Edn Engl. 56, 827–831 (2017).
Ember, S. W. et al. Acetyl-lysine binding site of bromodomain-containing protein 4 (BRD4) interacts with diverse kinase inhibitors. ACS Chem. Biol. 9, 1160–1171 (2014).
Ciceri, P. et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat. Chem. Biol. 10, 305–312 (2014).
Martin, M. P., Olesen, S. H., Georg, G. I. & Schönbrunn, E. Cyclin-dependent kinase inhibitor dinaciclib interacts with the acetyl-lysine recognition site of bromodomains. ACS Chem. Biol. 8, 2360–2365 (2013).
Erb, M. A. et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 543, 270–274 (2017).
Wan, L. et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 543, 265–269 (2017).
Bernard, D. et al. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene 24, 5543–5551 (2005).
Mohammad, H. P. et al. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 69, 6322–6330 (2009).
Perna, F. et al. Depletion of L3MBTL1 promotes the erythroid differentiation of human hematopoietic progenitor cells: possible role in 20q- polycythemia vera. Blood 116, 2812–2821 (2010).
Wang, J. X. et al. SPINDLIN1 promotes cancer cell proliferation through activation of WNT/TCF-4 signaling. Mol. Cancer Res. 10, 326–335 (2012).
Ren, C. et al. Structure-guided discovery of selective antagonists for the chromodomain of polycomb repressive protein CBX7. ACS Med. Chem. Lett. 7, 601–605 (2016).
Stuckey, J. I. et al. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat. Chem. Biol. 12, 180–187 (2016).
James, L. I. et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat. Chem. Biol. 9, 184–191 (2013).
James, L. I. et al. Small-molecule ligands of methyl-lysine binding proteins: optimization of selectivity for L3MBTL3. J. Med. Chem. 56, 7358–7371 (2013).
Robaa, D. et al. Identification and structure-activity relationship studies of small-molecule inhibitors of the methyllysine reader protein Spindlin1. ChemMedChem 11, 2327–2338 (2016).
Bae, N. et al. Developing Spindlin1 small-molecule inhibitors by using protein microarrays. Nat. Chem. Biol. 13, 750–756 (2017).
Shain, A. H. & Pollack, J. R. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8, e55119 (2013).
Anglesio, M. S. et al. Cancer-associated mutations in endometriosis without cancer. N. Engl. J. Med. 376, 1835–1848 (2017).
Guan, B. et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am. J. Surg. Pathol. 35, 625–632 (2011).
Helming, K. C. et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat. Med. 20, 251–254 (2014).
Hoffman, G. R. et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc. Natl Acad. Sci. USA 111, 3128–3133 (2014).
Wilson, B. G. et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol. Cell. Biol. 34, 1136–1144 (2014).
Lu, C. et al. Induction of sarcomas by mutant IDH2. Genes Dev. 27, 1986–1998 (2013).
Welch, J. S. et al. The origin and evolution of mutations in acute myeloid leukemia. Cell 150, 264–278 (2012).
Dang, L. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 (2009).
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008).
Xu, W. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011).
Chowdhury, R. et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463–469 (2011).
Sharma, H. Development of novel therapeutics targeting isocitrate dehydrogenase mutations in cancer. Curr. Top. Med. Chem. 18, 505–524 (2018).
Kim, E. S. Enasidenib: first global approval. Drugs 77, 1705–1711 (2017).
Stein, E. M. et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130, 722–731 (2017).
LaFave, L. M. et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat. Med. 21, 1344–1349 (2015).
Campbell, J. E. et al. EPZ011989, a potent, orally-available ezh2 inhibitor with robust in vivo activity. ACS Med. Chem. Lett. 6, 491–495 (2015).
Morschhauser, F. et al. Interim update from a phase 2 multicenter study of tazemetostat, an EZH2 inhibitor, in patients with relapsed or refractory (R/R) follicular lymphoma (FL). 23rd congress of the European Hematology Association. Stockholm: EHA Learning Center https://learningcenter.ehaweb.org/eha/2018/stockholm/21443/gilles.salles.interim.update.from.a.phase.2.multicenter.study.of.tazemetostat.html (2018).
Wyce, A. et al. MEK inhibitors overcome resistance to BET inhibition across a number of solid and hematologic cancers. Oncogenesis 7, 35 (2018).
Bradner, J. E., Hnisz, D. & Young, R. A. Transcriptional addiction in cancer. Cell 168, 629–643 (2017).
Kuendgen, A. et al. Efficacy of azacitidine is independent of molecular and clinical characteristics - an analysis of 128 patients with myelodysplastic syndromes or acute myeloid leukemia and a review of the literature. Oncotarget 9, 27882–27894 (2018).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21–29 (2015). 21 29.
Ponnaluri, V. K. C. et al. NicE-seq: high resolution open chromatin profiling. Genome Biol. 18, 122 (2017).
Broderick, J. M. FDA Halts enrollment on tazemetostat trials. OncLive https://www.onclive.com/web-exclusives/fda-halts-enrollment-on-tazemetostat-trials (2018).
Italiano, A. et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 19, 649–659 (2018).
Chandarlapaty, S. et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19, 58–71 (2011).
Duncan, J. S. et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012).
Zawistowski, J. S. et al. Enhancer remodeling during adaptive bypass to MEK inhibition is attenuated by pharmacologic targeting of the P-TEFb complex. Cancer Discov. 7, 302–321 (2017).
Nagarajan, S. et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 8, 460–469 (2014).
Feng, Q. et al. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res. 24, 809–819 (2014).
Asangani, I. A. et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282 (2014).
Asangani, I. A. et al. bet bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists in the treatment of prostate cancer. Mol. Cancer Res. 14, 324–331 (2016).
Xu, K. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–1469 (2012).
Knutson, S. K. et al. Synergistic anti-tumor activity of EZH2 inhibitors and glucocorticoid receptor agonists in models of germinal center non-hodgkin lymphomas. PLoS One 9, e111840 (2014).
Yang, X. P. et al. EZH2 is crucial for both differentiation of regulatory T cells and T effector cell expansion. Sci. Rep. 5, 10643 (2015).
Goswami, S. et al. Modulation of EZH2 expression in T cells improves efficacy of anti-CTLA-4 therapy. J. Clin. Invest. 128, 3813–3818 (2018).
Qin, Y. et al. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 38, 390–405 (2019).
Sheng, W. et al. LSD1 Ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563 e519 (2018).
Zhu, H. et al. BET bromodomain inhibition promotes anti-tumor immunity by suppressing PD-L1 expression. Cell Rep. 16, 2829–2837 (2016).
Harb, W. et al. A phase 1 study of CPI-1205, a small molecule inhibitor of EZH2, preliminary safety in patients with B-cell lymphomas. Ann. Oncol. 29, mdy048.001 (2018).
Maruyama, D. et al. First-in-human study of the EZH1/2 dual inhibitor DS-3201b in patients with relapsed or refractory non-Hodgkin lymphomas — preliminary results. Blood 130, 4070–4070 (2017).
Yap, T. A. et al. A phase I study of GSK2816126, an enhancer of zeste homolog 2(EZH2) inhibitor, in patients (pts) with relapsed/refractory diffuse large B-cell lymphoma (DLBCL), other non-hodgkin lymphomas (NHL), transformed follicular lymphoma (tFL), solid tumors and multiple myeloma (MM). Blood 128, 4203 (2016).
Shukla, N. et al. Final report of phase 1 study of the DOT1L inhibitor, pinometostat (EPZ-5676), in children with relapsed or refractory MLL-r acute leukemia. Blood 128, 2780 (2016).
O’Dwyer, P. J. et al. Abstract CT014: GSK525762, a selective bromodomain (BRD) and extra terminal protein (BET) inhibitor: results from part 1 of a phase I/II open-label single-agent study in patients with NUT midline carcinoma (NMC) and other cancers. Cancer Res. 76, CT014–CT014 (2016).
Somervaille, T. et al. Safety, phamacokinetics (PK), pharmacodynamics (PD) and preliminary activity in acute leukemia of Ory-1001, a first-in-class inhibitor of lysine-specific histone demethylase 1A (LSD1/KDM1A): initial results from a first-in-human phase 1 study. Blood 128, 4060–4060 (2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
All authors are employees and stockholders of GlaxoSmithKline, a pharmaceutical company that discovers and develops epigenetic therapies for cancer.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mohammad, H.P., Barbash, O. & Creasy, C.L. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med 25, 403–418 (2019). https://doi.org/10.1038/s41591-019-0376-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0376-8
This article is cited by
-
Cancer epigenetics: from laboratory studies and clinical trials to precision medicine
Cell Death Discovery (2024)
-
Targeting histone modifiers in bladder cancer therapy — preclinical and clinical evidence
Nature Reviews Urology (2024)
-
Proteomics analysis of histone deacetylase inhibitor-resistant solid tumors reveals resistant signatures and potential drug combinations
Acta Pharmacologica Sinica (2024)
-
Bridging tissue repair and epithelial carcinogenesis: epigenetic memory and field cancerization
Cell Death & Differentiation (2024)
-
A negative feedback loop between TET2 and leptin in adipocyte regulates body weight
Nature Communications (2024)