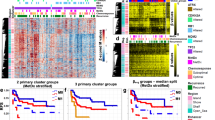
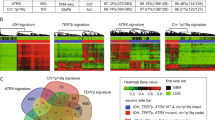
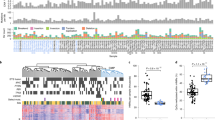
Abstract
Glioblastoma is characterized by widespread genetic and transcriptional heterogeneity, yet little is known about the role of the epigenome in glioblastoma disease progression. Here, we present genome-scale maps of DNA methylation in matched primary and recurring glioblastoma tumors, using data from a highly annotated clinical cohort that was selected through a national patient registry. We demonstrate the feasibility of DNA methylation mapping in a large set of routinely collected FFPE samples, and we validate bisulfite sequencing as a multipurpose assay that allowed us to infer a range of different genetic, epigenetic, and transcriptional characteristics of the profiled tumor samples. On the basis of these data, we identified subtle differences between primary and recurring tumors, links between DNA methylation and the tumor microenvironment, and an association of epigenetic tumor heterogeneity with patient survival. In summary, this study establishes an open resource for dissecting DNA methylation heterogeneity in a genetically diverse and heterogeneous cancer, and it demonstrates the feasibility of integrating epigenomics, radiology, and digital pathology for a national cohort, thereby leveraging existing samples and data collected as part of routine clinical practice.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ferlay, J. et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. International Agency for Research on Cancer http://globocan.iarc.fr (2013).
Woehrer, A., Bauchet, L. & Barnholtz-Sloan, J. S. Glioblastoma survival: has it improved? Evidence from population-based studies. Curr. Opin. Neurol. 27, 666–674 (2014).
Chinot, O. L. et al. Bevacizumab plus radiotherapy–temozolomide for newly diagnosed glioblastoma. N. Engl. J. Med. 370, 709–722 (2014).
Gilbert, M. R. et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N. Engl. J. Med. 370, 699–708 (2014).
Stupp, R. et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 15, 1100–1108 (2014).
Kim, H. et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 25, 316–327 (2015).
Kim, J. et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell 28, 318–328 (2015).
Kumar, A. et al. Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome Biol. 15, 530 (2014).
Lee, J. K. et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet. 49, 594–599 (2017).
Meyer, M. et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. USA 112, 851–856 (2015).
Patel, A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014).
Snuderl, M. et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 20, 810–817 (2011).
Sottoriva, A. et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 110, 4009–4014 (2013).
Wang, J. et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 48, 768–776 (2016).
Wang, Q. et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 33, 152 (2018).
Brennan, C. W. et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013).
Capper, D. et al. DNA methylation–based classification of central nervous system tumours. Nature 555, 469–474 (2018).
Ceccarelli, M. et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016).
Sturm, D. et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012).
Brocks, D. et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Reports 8, 798–806 (2014).
Mazor, T. et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell 28, 307–317 (2015).
Hao, J. J. et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 48, 1500–1507 (2016).
Lin, D. C. et al. Genomic and epigenomic heterogeneity of hepatocellular carcinoma. Cancer Res. 77, 2255–2265 (2017).
Li, S. et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat. Med. 22, 792–799 (2016).
Landau, D. A. et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26, 813–825 (2014).
Sheffield, N. C. et al. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 23, 386–395 (2017).
Wöhrer, A. et al. The Austrian Brain Tumour Registry: a cooperative way to establish a population-based brain tumour registry. J. Neurooncol. 95, 401–411 (2009).
Meissner, A. et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454, 766–770 (2008).
Klughammer, J. et al. Differential DNA methylation analysis without a reference genome. Cell Reports 13, 2621–2633 (2015).
Veillard, A. C., Datlinger, P., Laczik, M., Squazzo, S. & Bock, C. Diagenode® premium RRBS technology: cost-effective DNA methylation mapping with superior coverage. Nat. Methods 13, 184 (2016).
Bock, C. et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 28, 1106–1114 (2010).
Gu, H. et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat. Methods 7, 133–136 (2010).
Stefanits, H. et al. KINFix—a formalin-free noncommercial fixative optimized for histological, immunohistochemical and molecular analyses of neurosurgical tissue specimens. Clin. Neuropathol. 35, 3–12 (2016).
Bock, C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 13, 705–719 (2012).
Weller, M. et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat. Rev. Neurol. 6, 39–51 (2010).
Bienkowski, M. et al. Clinical Neuropathology practice guide 5-2015: MGMT methylation pyrosequencing in glioblastoma: unresolved issues and open questions. Clin. Neuropathol. 34, 250–257 (2015).
Mikeska, T. et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J. Mol. Diagn. 9, 368–381 (2007).
Turcan, S. et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012).
Wemmert, S. et al. Patients with high-grade gliomas harboring deletions of chromosomes 9p and 10q benefit from temozolomide treatment. Neoplasia 7, 883–893 (2005).
Verhaak, R. G. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010).
Bowman, R. L., Wang, Q., Carro, A., Verhaak, R. G. & Squatrito, M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro-oncol. 19, 139–141 (2017).
Sheffield, N. C. & Bock, C. LOLA: enrichment analysis for genomic region sets and regulatory elements in R and Bioconductor. Bioinformatics 32, (587–589 (2016).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Beier, C. P. et al. The cancer stem cell subtype determines immune infiltration of glioblastoma. Stem Cells Dev. 21, 2753–2761 (2012).
Strojnik, T. et al. Prognostic impact of CD68 and kallikrein 6 in human glioma. Anticancer Res. 29, 3269–3279 (2009).
Prosniak, M. et al. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin. Cancer Res. 19, 3776–3786 (2013).
Nowosielski, M. et al. Progression types after antiangiogenic therapy are related to outcome in recurrent glioblastoma. Neurology 82, 1684–1692 (2014).
Gentles, A. J. et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 131, 803–820 (2016).
Li, S. et al. Dynamic evolution of clonal epialleles revealed by methclone. Genome Biol. 15, 472 (2014).
Aldape, K. et al. GLASS Consortium. Glioma through the looking GLASS: molecular evolution of diffuse gliomas and the Glioma Longitudinal Analysis Consortium. Neuro-oncol. 20, 873–884 (2018).
Sahm, F. et al. DNA methylation–based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 18, 682–694 (2017).
McCord, M., Mukouyama, Y. S., Gilbert, M. R. & Jackson, S. Targeting WNT signaling for multifaceted glioblastoma therapy. Front. Cell. Neurosci. 11, 318 (2017).
Bock, C. et al. BLUEPRINT consortium. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat. Biotechnol. 34, 726–737 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Xi, Y. & Li, W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009).
Xi, Y. et al. RRBSMAP: a fast, accurate and user-friendly alignment tool for reduced representation bisulfite sequencing. Bioinformatics 28, 430–432 (2012).
Krueger, F. & Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, (1571–1572 (2011).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLOS Comput. Biol. 12, e1004873 (2016).
Kuilman, T. et al. CopywriteR: DNA copy number detection from off-target sequence data. Genome Biol. 16, 49 (2015).
Li, J. et al. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 17, 178–187 (2016).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Glaus, P., Honkela, A. & Rattray, M. Identifying differentially expressed transcripts from RNA-seq data with biological variation. Bioinformatics 28, 1721–1728 (2012).
Sahm, F. et al. Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 131, 903–910 (2016).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–7 (2016). W1.
Makambi, K. Weighted inverse chi-square method for correlated significance tests. J. Appl. Stat. 30, 225–234 (2003).
Assenov, Y. et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat. Methods 11, 1138–1140 (2014).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Hinrichs, A. S. et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34, D590–D598 (2006).
Lawson, J.T., Tomazou, E.M., Bock, C. & Sheffield, N.C. MIRA: an R package for DNA methylation-based inference of regulatory activity. Bioinformatics https://doi.org/10.1093/bioinformatics/bty083 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Deroulers, C. et al. Analyzing huge pathology images with open source software. Diagn. Pathol. 8, 92 (2013).
Schindelin, J., Rueden, C. T., Hiner, M. C. & Eliceiri, K. W. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 (2015).
Ruifrok, A. C. & Johnston, D. A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23, 291–299 (2001).
Phansalkar, N., More, S., Sabale, A. & Joshi, M. Adaptive local thresholding for detection of nuclei in diversity stained cytology images. In International Conference on Communications and Signal Processing (ICCSP) 218–220 (IEEE, 2011).
Vincent, L. & Soille, P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 13, 583–598 (1991).
Liu, Q. et al. Genetic, epigenetic, and molecular landscapes of multifocal and multicentric glioblastoma. Acta Neuropathol. 130, 587–597 (2015).
Porz, N. et al. Multi-modal glioblastoma segmentation: man versus machine. PLoS One 9, e96873 (2014).
Wen, P. Y. et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J. Clin. Oncol. 28, 1963–1972 (2010).
Nowosielski, M. et al. Radiologic progression of glioblastoma under therapy-an exploratory analysis of AVAglio. Neuro. Oncol. 20, 557–566 (2018).
Crammer, K. & Singer, Y. On the algorithmic implementation of multiclass kernel-based vector machines. J. Mach. Learn. Res. 2, 265–292 (2002).
Gentleman, R. & Temple Lang, D. Statistical analyses and reproducible research. Bioconductor project working papers. Working paper 2. http://biostats.bepress.com/bioconductor/paper2 (2004).
Acknowledgements
We thank all patients who have donated their samples for this study. We also thank G. Wilk, M. Muck, S. Schmid, and U. Andel for technical assistance with immunohistochemical stainings, macrodissection, and tumor tissue shavings; S. Mages for contributing to the interactive data visualization; the Biomedical Sequencing Facility at CeMM for assistance with next-generation sequencing; and all members of the Bock lab for their help and advice. The study was funded in part by an Austrian Science Fund grant (FWF KLI394) to A.W., a Marie Curie Career Integration Grant (European Union’s Seventh Framework Programme grant agreement no. PCIG12-GA-2012-333595) to C.B., an ERA-NET project grant (EpiMark FWF I 1575-B19) to C.B., an Austrian Science Fund grant (FWF I2714-B31) to G.L. and K.-H.N, and an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant agreement no. 640396) to B.B. Moreover, C.B. is supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant agreement no. 679146). Activities of the Austrian Brain Tumor Registry are supported by unrestricted research grants of Roche Austria to J.A.H. and the Austrian Society of Neurology to S.O. Some of the samples used for this research project were kindly provided by Biobank Graz.
Author information
Authors and Affiliations
Contributions
J. Klughammer, A.W., and C.B. designed the study. B.K., T.R., K.-H.N., J.F., N.P., M.N., M.A., M.M., T.S., G.L., B.B., J.A.H., and A.W. established and annotated the cohort. A.N. and P.D. performed DNA methylation profiling. D.A. performed low-coverage whole-genome sequencing. M.S. performed RNA-seq. J. Klughammer performed the data analysis. N.F., N.C.S, and B.E. contributed to data analysis. P.M., C.F.F., J. Kerschbaumer, C.T., A.E.G., G.S., M.K., S.O., F.M., S.W., J.T., J.B., J. Pichler, J.H., S.K., K.M.A., G.v.C., F.P., C.S., J. Preiser, T.H., P.A.W., W.K., F.W., T.B.-K., M.S., S.S., K.D., M.P., E.K., G.W., and C.M. contributed tumor samples and clinical data. J. Klughammer, A.W., and C.B. wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The optimized RRBS protocol that was used in this study has been licensed to Diagenode s.a. (Liège, Belgium) and commercialized as a kit and service.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13
Supplementary Table 1
Patient summary table
Supplementary Table 2
RRBS summary table
Supplementary Table 3
Survival analysis summary table
Supplementary Table 4
Association analysis summary table
Source Data Figure 1
Source Data Figure 1
Source Data Figure 2
Source Data Figure 2
Source Data Figure 3
Source Data Figure 3
Source Data Figure 4
Source Data Figure 4
Source Data Figure 5
Source Data Figure 5
Source Data Figure 6
Source Data Figure 6
Rights and permissions
About this article
Cite this article
Klughammer, J., Kiesel, B., Roetzer, T. et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med 24, 1611–1624 (2018). https://doi.org/10.1038/s41591-018-0156-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-018-0156-x
This article is cited by
-
DNA hypomethylator phenotype reprograms glutamatergic network in receptor tyrosine kinase gene-mutated glioblastoma
Acta Neuropathologica Communications (2024)
-
IDHwt glioblastomas can be stratified by their transcriptional response to standard treatment, with implications for targeted therapy
Genome Biology (2024)
-
Temporal change of DNA methylation subclasses between matched newly diagnosed and recurrent glioblastoma
Acta Neuropathologica (2024)
-
Compensatory cross-talk between autophagy and glycolysis regulates senescence and stemness in heterogeneous glioblastoma tumor subpopulations
Acta Neuropathologica Communications (2023)
-
Methylomics and cancer: the current state of methylation profiling and marker development for clinical care
Cancer Cell International (2023)