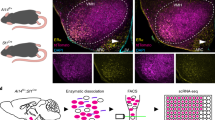
Abstract
Oestrogen depletion in rodents and humans leads to inactivity, fat accumulation and diabetes1,2, underscoring the conserved metabolic benefits of oestrogen that inevitably decrease with age. In rodents, the preovulatory surge in 17β-oestradiol (E2) temporarily increases energy expenditure to coordinate increased physical activity with peak sexual receptivity. Here we report that a subset of oestrogen-sensitive neurons in the ventrolateral ventromedial hypothalamic nucleus (VMHvl)3,4,5,6,7 projects to arousal centres in the hippocampus and hindbrain, and enables oestrogen to rebalance energy allocation in female mice. Surges in E2 increase melanocortin-4 receptor (MC4R) signalling in these VMHvl neurons by directly recruiting oestrogen receptor-α (ERα) to the Mc4r gene. Sedentary behaviour and obesity in oestrogen-depleted female mice were reversed after chemogenetic stimulation of VMHvl neurons expressing both MC4R and ERα. Similarly, a long-term increase in physical activity is observed after CRISPR-mediated activation of this node. These data extend the effect of MC4R signalling — the most common cause of monogenic human obesity8 — beyond the regulation of food intake and rationalize reported sex differences in melanocortin signalling, including greater disease severity of MC4R insufficiency in women9. This hormone-dependent node illuminates the power of oestrogen during the reproductive cycle in motivating behaviour and maintaining an active lifestyle in women.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mauvais-Jarvis, F., Clegg, D. J. & Hevener, A. L. The role of estrogens in control of energy balance and glucose homeostasis. Endocr. Rev. 34, 309–338 (2013).
Carr, M. C. The emergence of the metabolic syndrome with menopause. J. Clin. Endocrinol. Metab. 88, 2404–2411 (2003).
Correa, S. M. et al. An estrogen-responsive module in the ventromedial hypothalamus selectively drives sex-specific activity in females. Cell Rep. 10, 62–74 (2015).
Herber, C. B. et al. Estrogen signaling in arcuate Kiss1 neurons suppresses a sex-dependent female circuit promoting dense strong bones. Nat. Commun. 10, 163 (2019).
Martinez de Morentin, P. B. et al. Estradiol regulates brown adipose tissue thermogenesis via hypothalamic AMPK. Cell Metab. 20, 41–53 (2014).
Xu, Y. et al. Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab. 14, 453–465 (2011).
van Veen, J. E. et al. Hypothalamic estrogen receptor alpha establishes a sexually dimorphic regulatory node of energy expenditure. Nat. Metab. 2, 351–363 (2020).
Farooqi, I. S. et al. Dominant and recessive inheritance of morbid obesity associated with melanocortin 4 receptor deficiency. J. Clin. Invest. 106, 271–279 (2000).
Qi, L., Kraft, P., Hunter, D. J. & Hu, F. B. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum. Mol. Genet. 17, 3502–3508 (2008).
Thammacharoen, S., Lutz, T. A., Geary, N. & Asarian, L. Hindbrain administration of estradiol inhibits feeding and activates estrogen receptor-α-expressing cells in the nucleus tractus solitarius of ovariectomized rats. Endocrinology 149, 1609–1617 (2008).
Villanueva, E. C. et al. Complex regulation of mammalian target of rapamycin complex 1 in the basomedial hypothalamus by leptin and nutritional status. Endocrinology 150, 4541–4551 (2009).
Mountjoy, K. G., Mortrud, M. T., Low, M. J., Simerly, R. B. & Cone, R. D. Localization of the melanocortin-4 receptor (MC4-R) in neuroendocrine and autonomic control circuits in the brain. Mol. Endocrinol. 8, 1298–1308 (1994).
Ste Marie, L., Miura, G. I., Marsh, D. J., Yagaloff, K. & Palmiter, R. D. A metabolic defect promotes obesity in mice lacking melanocortin-4 receptors. Proc. Natl Acad. Sci. USA 97, 12339–12344 (2000).
Chen, A. S. et al. Role of the melanocortin-4 receptor in metabolic rate and food intake in mice. Transgenic Res. 9, 145–154 (2000).
Huszar, D. et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88, 131–141 (1997).
Sina, M. et al. Phenotypes in three pedigrees with autosomal dominant obesity caused by haploinsufficiency mutations in the melanocortin-4 receptor gene. Am. J. Hum. Genet. 65, 1501–1507 (1999).
Skene, P. J., Henikoff, J. G. & Henikoff, S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018).
Lin, C. Y. et al. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet. 3, e87 (2007).
Lo, L. et al. Connectional architecture of a mouse hypothalamic circuit node controlling social behavior. Proc. Natl Acad. Sci. USA 116, 7503–7512 (2019).
Fuhrmann, F. et al. Locomotion, theta oscillations, and the speed-correlated firing of hippocampal neurons are controlled by a medial septal glutamatergic circuit. Neuron 86, 1253–1264 (2015).
Góis, Z. H. T. D. & Tort, A. B. L. Characterizing speed cells in the rat hippocampus. Cell Rep. 25, 1872–1884 (2018).
Evans, D. A. et al. A synaptic threshold mechanism for computing escape decisions. Nature 558, 590–594 (2018).
Tovote, P. et al. Midbrain circuits for defensive behaviour. Nature 534, 206–212 (2016).
Carter, M. E. et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat. Neurosci. 13, 1526–1533 (2010).
Coimbra, B. et al. Role of laterodorsal tegmentum projections to nucleus accumbens in reward-related behaviors. Nat. Commun. 10, 4138 (2019).
Balthasar, N. et al. Divergence of melanocortin pathways in the control of food intake and energy expenditure. Cell 123, 493–505 (2005).
Matharu, N. et al. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363, eaau0629 (2019).
Slonaker, J. R. The effect of pubescence, oestruation and menopause on the voluntary activity in the albino rat. Am. J. Physiol. 68, 294–315 (1924).
Lotta, L. A. et al. Human gain-of-function MC4R variants show signaling bias and protect against obesity. Cell 177, 597–607 (2019).
Pfaus, J. G., Shadiack, A., Van Soest, T., Tse, M. & Molinoff, P. Selective facilitation of sexual solicitation in the female rat by a melanocortin receptor agonist. Proc. Natl Acad. Sci. USA 101, 10201–10204 (2004).
Clayton, A. H. et al. Bremelanotide for female sexual dysfunctions in premenopausal women: a randomized, placebo-controlled dose-finding trial. Womens Health 12, 325–337 (2016).
Chandler, D. J. et al. Redefining noradrenergic neuromodulation of behavior: impacts of a modular locus coeruleus architecture. J. Neurosci. 39, 8239–8249 (2019).
Duval, K. et al. Effects of the menopausal transition on energy expenditure: a MONET Group Study. Eur. J. Clin. Nutr. 67, 407–411 (2013).
O’Neal, T. J., Friend, D. M., Guo, J., Hall, K. D. & Kravitz, A. V. Increases in physical activity result in diminishing increments in daily energy expenditure in mice. Curr. Biol. 27, 423–430 (2017).
Duggal, N. A., Niemiro, G., Harridge, S. D. R., Simpson, R. J. & Lord, J. M. Can physical activity ameliorate immunosenescence and thereby reduce age-related multi-morbidity? Nat. Rev. Immunol. 19, 563–572 (2019).
Garfield, A. S. et al. A neural basis for melanocortin-4 receptor-regulated appetite. Nat. Neurosci. 18, 863–871 (2015).
Dhillon, H. et al. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron 49, 191–203 (2006).
Krashes, M. J. et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011).
Cai, D., Cohen, K. B., Luo, T., Lichtman, J. W. & Sanes, J. R. Improved tools for the Brainbow toolbox. Nat. Methods 10, 540–547 (2013).
Hu, Y. et al. Differential effects of unfolded protein response pathways on axon injury-induced death of retinal ganglion cells. Neuron 73, 445–452 (2012).
Nagai, Y. et al. Deschloroclozapine, a potent and selective chemogenetic actuator enables rapid neuronal and behavioral modulations in mice and monkeys. Nat. Neurosci. 23, 1157–1167 (2020).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137.
Hahne, F. & Ivanek, R. Visualizing genomic data using Gviz and Bioconductor. Methods Mol. Biol. 1418, 335–351 (2016).
Crane, J. D., Mottillo, E. P., Farncombe, T. H., Morrison, K. M. & Steinberg, G. R. A standardized infrared imaging technique that specifically detects UCP1-mediated thermogenesis in vivo. Mol Metab. 3, 490–494 (2014).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002).
Acknowledgements
We thank C. Paillart and T. McMahon for technical assistance with the running and data acquisition for the CLAMS and ANY-maze systems; all members of the Ingraham laboratory for their comments and discussions; O. Yabut for expertise in imaging; and C. Vaisse for insights on MC4R signalling. This work was supported by funding to H.A.I. (R01DK121657, R01AG062331, UCSF Women’s Reproductive Health RAP Award and GCRLE Senior Scholar Award 0320), to W.C.K. (American Heart Association Postdoctoral Fellowship 16POST27260361), to R.R. (K12GM081266 IRACDA Program), to B.G. (2T32GM065094, F31MH124365), to N.M. (UCSF Mary Ann Koda-Kimble Innovation Seed Award, UCSF Catalyst Program), to X.D. (R01EY030138, Research to Prevent Blindness and Klingenstein‐Simons Neuroscience Fellowship), to S.M.C. (K01 DK098320, UCLA Women’s Health Center, UL1TR001881), to C.B.H. (F32 DK107115-01A1, AHA Postdoctoral Fellowship 16POST29870011), to N.A. (R01CA197139, R01 MH109907), to J.T. (R01MH113628, SFARI600568). We acknowledge the mouse metabolic core funded by P30 DK098722-01.
Author information
Authors and Affiliations
Contributions
W.C.K. designed experiments, analysed data and wrote the manuscript. R.R. performed thermal and glucose homeostasis analyses in mice. B.G. optimized, performed and analysed the CUT&RUN method for ERα binding in neurons. N.M. provided CRISPRa viral vectors and expert advice. A.N.R. performed histology experiments and quantification of expression data. A.M.P.-R. aided with chemogenetic data acquisition and analyses. C.B.H. analysed bone and plasma lipid data. S.M.C. designed experiments, provided animal models and analysed data. K.T. and X.D. provided the AAV-DIO-mYFP vector. N.A. provided key unpublished reagents related to CRISPRa constructs and helped to guide studies. J.T. optimized CUT&RUN method for ERα binding in neurons, performed analyses and wrote the manuscript. H.A.I. designed experiments, analysed data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Miguel López and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 VMHvlERαKO does not affect energy intake or physical activity during the light phase but does decrease iBAT thermogenic gene expression.
a, Representative image of a successful hit confirmed post-mortem by loss of ERα expression (red) in the VMHvl (white arrow) with corresponding expression of GFP driven by the AAV2-Cre viral vector. Scale bars, 100 µm. b, Food intake and X-ambulatory parameters obtained in light period for VMHvlErαKO (n = 10) and ARCERαKO (n = 12) female cohorts compared to controls (n = 7 and 5). c, d, Equivalent fat mass in VMHvlERαKO (n = 9), ARCERαKO (n = 11), and control (n = 7 and 6) (c) and oxygen consumption rates in VMHvlERαKO (n = 12), ARCERαKO (n = 11), and control (n = 6 and 6) (d) female mice measured by EchoMRI and indirect calorimetry in CLAMS, respectively. e, Quantification of BAT thermogenic gene expression levels by qPCR in VMHvlERαKO (n = 6), ARCERαKO (n = 7), and control (n = 6 and 3) female mice (VMHvlERαKO Ucp1: unpaired two-tailed Student's t-test, t9 = 2.599, P = 0.0288).
Extended Data Fig. 2 Induction of pS6 and Mc4r in the VMHvl depends on oestrogen signalling through ERα.
a, Immunofluorescence microscopy images of ERα and pS6(S244/S247) in the VMHvl (left panels) and ARC (right panels) of Esr1fl/fl and OVX female mice 4 h post oestradiol benzoate treatment. b, Immunofluorescence microscopy images of ERα (red) and pS6(S244/S247) (green) in the VMHvl in conditional knockout (Esr1Nkx2-1Cre) 4OVX female mice following 4 h after oestradiol benzoate treatment. c, Immunofluorescence microscopy images of ERα (red) and pS6(S244/S247) (green) in the VMHvl of intact adult male mice following 4 h post vehicle or oestradiol benzoate treatment. Bar graph to the right shows increased number of pS6(S244/S247)-positive neurons in the VMHvl of male mice after oestradiol benzoate treatment as done for OVX female mice (unpaired two-tailed t-test, t6 = 8.569, P = 0.0001). d, Mammalian Phenotype Ontology terms most significantly enriched and top five significantly enriched Reactome Pathways among DEGs in the VMHvl (vehicle versus oestradiol benzoate). e, qPCR analysis of the indicated target genes in VMHvl from oestrus female mice, proestrus female mice, and male mice; data points represent values from individual mice (one-way ANOVA Nmur2: F2,15 = 8.469, P = 0.0035, post hoc: oestrus (E) versus proestrus (P) P = 0.0454, proestrus versus male P = 0.0030, and oestrus versus male P = 0.1438; Esr1: F2,11 = 10.18, P = 0.0031, post hoc: oestrus versus proestrus P = 0.1650, proestrus versus male P = 0.0374, and oestrus versus male P = 0.0033). Holm–Šidák multiple comparisons test. f, Mc4r expression levels in VMHvl from OVX oestradiol-benzoate-treated (n = 6) female mice normalized to vehicle treatment (n = 3) (unpaired two-tailed Student's t-test t6 = 6.519, P = 0.0006). g, ISH showing Mc4r expression (red arrows) in the VMHvl of an oestrus female, a proestrus female, and an intact male. h, Representative ISH (Mc4r, red arrows) and immunofluorescent (pS6, yellow arrows) staining in the VMHvl (dashed circle) from OVX female mice treated with vehicle for 4 h, oestradiol benzoate for 2 h, or oestradiol benzoate for 4 h. i, Full size images showing bilateral expression of Mc4r and Rprm in intact female mice staged for oestrus and proestrus. Rprm expression is unchanged in both oestrous stages. Data presented as box plots (see Fig. 1c legend for description). Micrographs are representative of data from 5 mice.
Extended Data Fig. 3 ERα-binding sites in oestradiol-benzoate-sensitive target genes contain conserved ERE consensus sequences.
CUT&RUN CPM-normalized coverage track showing oestradiol-benzoate-specific ERα binding sites containing EREs (pink boxes) within the Greb1 locus (a; 3/3 replicates) and Pgr locus (b; 3/3 replicates), and in the Mc4r promoter (c; 1 of 3 replicates) in 400,000 sub-cortical brain nuclei collected from vehicle and oestradiol benzoate (5 µg) treated gonadectomized mice. Below each track the location and sequence conservation of full (a, b) ERE and half (c) SP1/ERE consensus sites in target gene loci indicated by pink and green boxes. d, e, Location and sequence conservation of ERE consensus sites within Mc4r and Nmur2 loci corresponding to ERα binding sites presented in Fig 1g. For all panels the genomic intervals containing ERE/SP1 sites are located within the ERα binding sites identified by CUT&RUN.
Extended Data Fig. 4 Limited induction of Mc4r expression in the MeA by ERα signalling, and additional neuroanatomical targets of VMHvlMC4R neurons.
a, Representative coronal brain images of Ai14Mc4r female mice stained for ERα (green) and Ai14 (magenta) in the MeA used for quantification shown in Fig 2a. b, Additional ISH comparing Mc4r induction in the VMHvl and MEA in oestrus and proestrus female mice. c, Representative mYFP reporter expression in additional neuroanatomical regions. Scale bars, 200 µm. d, Heat map from Fig. 2d rearranged to compare VMHvlMC4R and VMHvlERα projection intensity in target regions along the anterior-posterior axis. e, VMHvlMC4R projections to the PAG preferentially target the PAGdl/l and PAGdm. Scale bars, 200 µm.
Extended Data Fig. 5 VMHvlMC4R neurons function specifically to drive physical activity.
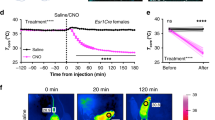
a, Distance travelled over time in female and male mice following a single injection of CNO (repeated-measures two-way ANOVA interaction effect female: F39,312 = 11.96, P < 0.0001; male: F39,312 = 6.898, P < 0.0001). Holm–Šidák multiple comparisons test. b, Total time spent immobile in intact female and male VMHvlCre− controls (n = 5/4) and VMHvlMC4R::hM3Dq mice (n = 5/5) (repeated-measures two-way ANOVA female interaction effect F1,8 = 33.89, P = 0.0004, post hoc P < 0.0001 and male interaction effect F1,8 = 96.79, P = 0.0005, post hoc P < 0.0001). Holm–Šidák multiple comparisons test. c, Thermal imaging of BAT surface temperatures for VMHvlCre− (left mouse) and VMHvlMC4R:: hM3Dq (right mouse) female mice treated with CL-316,243. d, No differences were observed in Ucp1 mRNA in the BAT from VMHvlCre− and VMHvlMC4R::hM3Dq mice collected 1.5 h after a single CNO injection. e, Body weights for female VMHvlCre− (n = 5) and VMHvlMC4R::hM3Dq (n = 6, baseline measurement includes 1 mouse with mistargeted injection) mice prior to glucose tolerance test (GTT). f, GTT glucose levels for intact female cohorts treated with saline or CNO and total area under the curve (AUC) (unpaired two-tailed Student's t-test t8 = 2.824, P = 0.0223). g, Body weight normalized food consumption in female mice (n = 5/5) following Sal/CNO injection during light dark period (ZT12–ZT17) (repeated-measures two-way ANOVA Dark period interaction effect F1,8 = 3.502, P = 0.0982, post hoc P = 0.0489). Holm–Šidák multiple comparisons test. h, Sustained physical activity increase across light/dark periods in VMHvlMC4R::hM3Dq (n = 5) female mice administered CNO-H2O as compared to VMHvlCre− female mice (n = 5) or during exposure to plain drinking water (H2O). i, j, Cumulative distance travelled (i) and number of rearing events (j) during light/dark periods following administration of CNO or water during the light stage (i repeated-measures two-way ANOVA F1,8 = 15.8, P = 0.0041, post hoc P = 0.0006 and j, repeated-measures two-way ANOVA F1,8 = 15.8, P = 0.0041, post hoc P = 0.0006). Holm–Šidák multiple comparisons test. k, Starting body weights and weight change during continuous administration of CNO-H2O for intact female mice. l, ERα and mCherry expression in the VMHvl following targeted injection of Cre-dependent AAV-hM4Di-mCherry into a female Mc4r-t2a-cre mouse. m, Number of rearing episodes during the dark period in VMHvlMC4R::hM4Di (n = 8) and VMHvlCre− (n = 4) intact female mice following administration of plain or DCZ-laden drinking water. Data are mean ± s.e.m. or box plots (described in Fig. 1c legend).
Extended Data Fig. 6 Increased physical activity and improvement in metabolic health markers in OVX VMHvlMC4R::hM3Dq female mice in response to acute and chronic CNO.
a, Distance travelled over time in OVX female VMHvlCre− (n = 5) and VMHvlMC4R::hM3Dq (n = 5) mice during administration of plain H2O or CNO-H2O (repeated-measures two-way ANOVA interaction effect F11,88 = 5.265, P < 0.0001). b, Total dark period rearing events in intact and OVX female mice administered plain H2O or CNO-H2O drinking water (repeated-measures two-way ANOVA interaction effect F1,8 = 60.31, P < 0.0001 post hoc P < 0.0001). c, Body weights (repeated-measures two-way ANOVA time effect F2,24 = 49.51, P < 0.0001; genotype effect F2,24 = 33.50, P < 0.0001) and fasting glucose levels (repeated-measures two-way ANOVA time effect F2,26 = 6.456, P = 0.0053; genotype effect F1,26 = 10.11, P = 0.0038) in female mice after OVX and subsequent HFD feeding. d, Blood glucose (left) and AUC (right) during ITT in chow-fed OVX female mice following 6-hour fast and saline/CNO treatment. e, Blood glucose (left, repeated-measures two-way ANOVA interaction effect F1,8 = 7.791, P = 0.0235, post hoc: VMHvlMC4R::hM3Dq saline versus CNO, T15 P = 0.0009 and T60 P = 0.0318) and AUC (right, repeated-measures ANOVA with mixed-effects model, note: one cre+ female with missed injection was included in saline but not CNO treated groups, interaction effect F1,8 = 7.791, P = 0.0235, post hoc: VMHvlMC4R::hM3Dq saline versus CNO P = 0.0007) values during 90 min ITT test on HFD-fed OVX female mice performed 6 hours post fasting and post injection with saline or CNO. f, Blood glucose levels following a 6 h fast in OVX female mice maintained on Chow/HFD following a single saline or CNO injection (repeated-measures ANOVA with mixed-effects model, note: one cre+ female with missed injection was included in saline but not CNO treated groups, Chow: treatment effect F1,17 = 5.038, P = 0.0384, post hoc P = 0.0179; and HFD: interaction effect F1,17 = 20.47, P = 0.0019, post hoc P = 0.0073). g, Percentage change in body weight in HFD-fed OVX female mice (n = 5/5) continuously administered CNO-laden drinking water (repeated-measures two-way ANOVA interaction effect F7,64 = 4.583, P = 0.0003. h, Fasting blood glucose levels in OVX/HFD mice before and after 8 days of chronic CNO (repeated-measures ANOVA with mixed-effects model, note: one cre+ female with missed injection was included in saline but not CNO treated groups, interaction effect F1,17 = 5.180, P = 0.0361 post hoc: VMHvlMC4R::hM3Dq Pre versus Post P = 0.0156). i, Plasma cholesterol levels before (Pre) and after (Post) 8 days of continuous CNO-H2O exposure (repeated-measures ANOVA with mixed-effects model, note: one cre+ female with missed injection was included in saline- but not CNO-treated groups, interaction effect F1,8 = 5.502, P = 0.0470, post hoc P = 0.0203). j, Average daily food intake during 8 days of continuous CNO-H2O exposure (points represent separate daily measurements of average consumption per mouse). Data are mean ± s.e.m. or box plots (described in Fig. 1c legend). a–h, ANOVA with Holm–Šidák multiple comparisons test.
Extended Data Fig. 7 Additional metabolic and expression data for conditional Mc4r-rescue mice.
a, FOS expression (arrows) in the VMHvl and PVH of female mice treated with oestradiol benzoate ± MT-II. b, FOS+ cells in oestradiol benzoate + MT-II (n = 5) compared to vehicle (Veh) + MT-II (n = 3, **P = 0.0037) and oestradiol benzoate + vehicle (n = 4, **P = 0.0046) treated female mice (one-way ANOVA F2,9 = 14.00, P = 0.0017). c, Mc4r and Sf1 expression patterns from Genotype-Tissue Expression Project intersect specifically in the hypothalamus (blue arrows) and not in peripheral tissues (red arrows). Transcripts/million (TPM) expression presented as box plots with centre line at median, box edges are 25th and 75th percentiles, and whiskers are 1.5x interquartile range. d, Equivalent body weights within cohorts of female and male Mc4r+/+, Mc4rloxTB and Mc4rSf1-cre mice at weaning. e, Percentage lean (one-way ANOVA F2,31 = 101.4, P < 0.0001, post hoc: Mc4r+/+ versus Mc4rloxTB P < 0.0001, Mc4r+/+ versus Mc4rSf1-cre P < 0.0001, and Mc4rSf1-cre versus Mc4rloxTB P = 0.0720) and % fat (one-way ANOVA F2,31 = 104.2, P < 0.0001, post hoc: Mc4r+/+ versus Mc4rloxTB P < 0.0001, Mc4r+/+ versus Mc4rSf1-cre P < 0.0001, and Mc4rSf1-cre versus Mc4rloxTB P=0.0769) body composition analysis (EchoMRI) in adult female mice of each genotype. f, Oxygen consumption (VO2) as a function of body weight in adult female mice. g, Body weights in 13-week-old Mc4r+/+, Mc4rloxTB, and Mc4rSf1-cre female mice (one-way ANOVA F2,32 = 226.6, post hoc: Mc4r+/+ versus Mc4rloxTB P < 0.0001, Mc4r+/+ versus Mc4rSf1-cre P < 0.0001, and Mc4rSf1-cre versus Mc4rloxTB P=0.0029). Data presented as mean ± s.e.m. or scatterplots of values from individual mice. Number of mice analysed are indicated on each bar. b, d-g, Holm–Šidák multiple comparisons test.
Extended Data Fig. 8 Expression and physical activity levels in male and female Mc4rCRISPRa mice.
a, mCherry expression in Mc4rCRISPRa female hypothalamus. b, Fluorescent ISH images from Mc4rCRISPRa female (left) and male (right) showing Esr1 and Mc4r expression. Images are reproduced and expanded from Fig. 4g to show limited induction of Mc4r outside of the VMHvl target region. c, Dark phase (ZT12–ZT24) physical activity levels (distance per 12 h) as a function of Mc4r or mCherry mRNA expression in microdissected VMHvl from control and Mc4rCRISPRa female mice. d, Home-cage activity in Mc4rCRISPRa (n = 4) and control (n = 3) male mice. e, Time spent immobile during the 12 hour dark phase in control and Mc4rCRISPRa female (unpaired two-tailed Student's t-test, t9 = 2.015, P = 0.0747) and male Mc4rCRISPRa mice (see Fig. 4 for number of mice per group) (unpaired two-tailed Student's t-test, t5 = 3.245, P = 0.0228). f, Mc4rCRISPRa (n = 6) and control (n = 7) female body weights during ad lib feeding. g, Normalized daily food intake in Mc4rCRISPRa (n = 6) and control (n = 7) female mice (unpaired two-tailed Student's t-test, t11 = 2.409, *P = 0.0347). h, BAT surface temperatures in female control (n = 4) and Mc4rCRISPRa (n = 5) mice, repeated measurements at 30- and 60-min post-anaesthesia. i, Cortical bone thickness for female cohorts (unpaired two-tailed t-test, t6 = 2.957, P = 0.0254). j, Body weights in control (n = 4) and Mc4rCRISPRa (n = 6) female mice at wk 9 and at wk 17 after eight weeks of OVX. k, Distance travelled over 24 hours in OVX control and OVX Mc4rCRISPRa compared to intact female mice (blue). l, Total dark phase distance in intact (n = 5), OVX control (n = 7), and OVX Mc4rCRISPRa (n = 8) female mice. Data are presented as scatterplots of values from individual mice, mean ± s.e.m., or as box plots (described in the legend of Fig. 1c).
Supplementary information
Supplementary Video 1
CNO administered in the drinking water stimulates VMHvlMC4R neurons to increase physical activity. Video recording of VMHvlMC4R::hM3dq (top) and VMHvlCre− (bottom) female mice following addition of CNO-laden drinking water (0.25 mg ml−1) during the inactive, lights-on period. Recordings have been sped up 20×.
Rights and permissions
About this article
Cite this article
Krause, W.C., Rodriguez, R., Gegenhuber, B. et al. Oestrogen engages brain MC4R signalling to drive physical activity in female mice. Nature 599, 131–135 (2021). https://doi.org/10.1038/s41586-021-04010-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04010-3
This article is cited by
-
Artificial ovaries constructed from biodegradable chitin-based hydrogels with the ability to restore ovarian endocrine function and alleviate osteoporosis in ovariectomized mice
Reproductive Biology and Endocrinology (2023)
-
Chemogenetics for cell-type-specific modulation of signalling and neuronal activity
Nature Reviews Methods Primers (2023)
-
The ventromedial hypothalamic nucleus: watchdog of whole-body glucose homeostasis
Cell & Bioscience (2022)
-
Sex-specific multi-level 3D genome dynamics in the mouse brain
Nature Communications (2022)
-
Gene regulation by gonadal hormone receptors underlies brain sex differences
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.