Abstract
The initiation of transcription is a focal point for the regulation of gene activity during mammalian cell differentiation and development. To initiate transcription, RNA polymerase II (Pol II) assembles with general transcription factors into a pre-initiation complex (PIC) that opens promoter DNA. Previous work provided the molecular architecture of the yeast1,2,3,4,5,6,7,8,9 and human10,11 PIC and a topological model for DNA opening by the general transcription factor TFIIH12,13,14. Here we report the high-resolution cryo-electron microscopy structure of PIC comprising human general factors and Sus scrofa domesticus Pol II, which is 99.9% identical to human Pol II. We determine the structures of PIC with closed and opened promoter DNA at 2.5–2.8 Å resolution, and resolve the structure of TFIIH at 2.9–4.0 Å resolution. We capture the TFIIH translocase XPB in the pre- and post-translocation states, and show that XPB induces and propagates a DNA twist to initiate the opening of DNA approximately 30 base pairs downstream of the TATA box. We also provide evidence that DNA opening occurs in two steps and leads to the detachment of TFIIH from the core PIC, which may stop DNA twisting and enable RNA chain initiation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM density reconstructions and final models have been deposited with the Electron Microscopy Data Bank (EMDB) (accession codes EMD-12611–EMD-12630) and with the Protein Data Bank (PDB) (proximal cPIC: 7NVS, distal cPIC: 7NVT, TFIIH (CC): 7NVW, cPIC (OC): 7NVU, TFIIH (OC): 7NVX, XPB (intermediary complex): 7NVV, proximal CC: 7NVY, distal CC: 7NVZ, OC: 7NW0). All data are available in the Article or its Supplementary Information files. Publicly available PDB entries used in this study are: 5FLM, 5IY7, 5GPY, 6NMI and 6RO4.
References
Kostrewa, D. et al. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009).
Sainsbury, S., Niesser, J. & Cramer, P. Structure and function of the initially transcribing RNA polymerase II–TFIIB complex. Nature 493, 437–440 (2013).
Plaschka, C. et al. Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016).
Plaschka, C. et al. Architecture of the RNA polymerase II–Mediator core initiation complex. Nature 518, 376–380 (2015).
Dienemann, C., Schwalb, B., Schilbach, S. & Cramer, P. Promoter distortion and opening in the RNA polymerase II cleft. Mol. Cell 73, 97–106 (2019).
Schilbach, S. et al. Structures of transcription pre-initiation complex with TFIIH and Mediator. Nature 551, 204–209 (2017).
Liu, X., Bushnell, D. A., Wang, D., Calero, G. & Kornberg, R. D. Structure of an RNA polymerase II–TFIIB complex and the transcription initiation mechanism. Science 327, 206–209 (2010).
Robinson, P. J. et al. Structure of a complete mediator–RNA polymerase II pre-initiation complex. Cell 166, 1411–1422 (2016).
Murakami, K. et al. Structure of an RNA polymerase II preinitiation complex. Proc. Natl Acad. Sci. USA 112, 13543–13548 (2015).
He, Y., Fang, J., Taatjes, D. J. & Nogales, E. Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013).
He, Y. et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016).
Sainsbury, S., Bernecky, C. & Cramer, P. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143 (2015).
Hahn, S. & Young, E. T. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189, 705–736 (2011).
Fishburn, J., Tomko, E., Galburt, E. & Hahn, S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc. Natl Acad. Sci. USA 112, 3961–3966 (2015).
Nogales, E., Louder, R. K. & He, Y. Structural insights into the eukaryotic transcription initiation machinery. Annu. Rev. Biophys. 46, 59–83 (2017).
Hantsche, M. & Cramer, P. Conserved RNA polymerase II initiation complex structure. Curr. Opin. Struct. Biol. 47, 17–22 (2017).
Mühlbacher, W. et al. Conserved architecture of the core RNA polymerase II initiation complex. Nat. Commun. 5, 4310 (2014).
Grünberg, S., Warfield, L. & Hahn, S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat. Struct. Mol. Biol. 19, 788–796 (2012).
Kim, T. K., Ebright, R. H. & Reinberg, D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288, 1418–1422 (2000).
Greber, B. J. et al. The cryo-electron microscopy structure of human transcription factor IIH. Nature 549, 414–417 (2017).
Greber, B. J., Toso, D. B., Fang, J. & Nogales, E. The complete structure of the human TFIIH core complex. eLife 8, e44771 (2019).
Kokic, G. et al. Structural basis of TFIIH activation for nucleotide excision repair. Nat. Commun. 10, 2885 (2019).
Compe, E., Genes, C. M., Braun, C., Coin, F. & Egly, J. M. TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription. Nat. Commun. 10, 2084 (2019).
Farnung, L., Ochmann, M. & Cramer, P. Nucleosome-CHD4 chromatin remodeler structure maps human disease mutations. eLife 9, e56178 (2020).
Li, M. et al. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 567, 409–413 (2019).
Wang, W., Carey, M. & Gralla, J. D. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science 255, 450–453 (1992).
Holstege, F. C., van der Vliet, P. C. & Timmers, H. T. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15, 1666–1677 (1996).
Giardina, C. & Lis, J. T. DNA melting on yeast RNA polymerase II promoters. Science 261, 759–762 (1993).
Tomko, E. J., Fishburn, J., Hahn, S. & Galburt, E. A. TFIIH generates a six-base-pair open complex during RNAP II transcription initiation and start-site scanning. Nat. Struct. Mol. Biol. 24, 1139–1145 (2017).
Ren, D., Lei, L. & Burton, Z. F. A region within the RAP74 subunit of human transcription factor IIF is critical for initiation but dispensable for complex assembly. Mol. Cell. Biol. 19, 7377–7387 (1999).
Tomko, E. J. et al. The role of XPB/Ssl2 dsDNA translocase processivity in transcription start-site scanning. J. Mol. Biol. https://doi.org/10.1016/j.jmb.2021.166813 (2021).
Qiu, C. et al. Universal promoter scanning by Pol II during transcription initiation in Saccharomyces cerevisiae. Genome Biol. 21, 132 (2020).
Murakami, K. et al. Uncoupling promoter opening from start-site scanning. Mol. Cell 59, 133–138 (2015).
Rengachari, S., Schilbach, S., Aibara, S., Dienemann, C. & Cramer, P. Structure of the human Mediator–RNA polymerase II pre-initiation complex. Nature https://doi.org/10.1038/s41586-021-3555-7 (2021).
Gradia, S. D. et al. MacroBac: new technologies for robust and efficient large-scale production of recombinant multiprotein complexes. Methods Enzymol. 592, 1–26 (2017).
Berrow, N. S. et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 35, e45 (2007).
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Vos, S. M., Farnung, L., Urlaub, H. & Cramer, P. Structure of paused transcription complex Pol II–DSIF–NELF. Nature 560, 601–606 (2018).
Hu, X. et al. A Mediator-responsive form of metazoan RNA polymerase II. Proc. Natl Acad. Sci. USA 103, 9506–9511 (2006).
Kastner, B. et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2008).
Seizl, M., Larivière, L., Pfaffeneder, T., Wenzeck, L. & Cramer, P. Mediator head subcomplex Med11/22 contains a common helix bundle building block with a specific function in transcription initiation complex stabilization. Nucleic Acids Res. 39, 6291–6304 (2011).
Noe Gonzalez, M., Sato, S., Tomomori-Sato, C., Conaway, J. W. & Conaway, R. C. CTD-dependent and -independent mechanisms govern co-transcriptional capping of Pol II transcripts. Nat. Commun. 9, 3392 (2018).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019).
Scheres, S. H. W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Acknowledgements
We thank members of the Cramer laboratory, in particular C. Dienemann and U. Steuerwald for maintenance of the cryo-EM facility, U. Neef and P. Rus for maintenance of the insect cell facility, and F. Jochheim and M. Klein for maintenance of the computing infrastructure. S.A. was supported by an H2020 Marie Curie Individual Fellowship (894862). P.C. was supported by the Deutsche Forschungsgemeinschaft (EXC 2067/1 39072994, SFB860, SPP2191) and the ERC Advanced Investigator Grant CHROMATRANS (grant agreement no. 882357).
Author information
Authors and Affiliations
Contributions
S.A. designed and carried out data analysis and model building. S.S. designed and carried out experiments. P.C. designed and supervised research. S.A., S.S. and P.C. interpreted the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Steve Hahn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
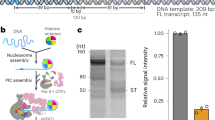
Extended Data Fig. 1 Biochemical assembly of PIC samples, and PIC cryo-EM maps.
a, SDS–PAGE analysis of S. scrofa domesticus Pol II, H. sapiens general transcription factors and a peak fraction of mammalian PIC assembled on a sucrose gradient (replicated three times). b, Representative cryo-EM micrograph. Left, without ADP·BeF3 (replicated 30,110 times). Right, with ADP·BeF3 (replicated 67,108 times). Scale bar, 300 Å. c, d, Cryo-EM reconstructions used for model building of the CC-states (without ADP·BeF3) (c) and OC-state (with ADP·BeF3) (d). The resolution for the respective focused refined maps is provided in parentheses, and an outline of the lower-resolution map used to dock in the focused maps is shown. e, Promoter-dependent transcription assay demonstrates that our PIC preparation is initiation competent. Asterisk indicates expected RNA product length. For the negative control, TFIIE and TFIIH were lacking from the reaction (replicated three times).
Extended Data Fig. 2 Processing tree of cryo-EM dataset of PIC without ADP·BeF3.
Processing tree indicating how the particles were assigned their classes and angles for this study. Reconstructions with an orange backing indicate high-resolution maps used for initial building, which were then fitted into reconstructions with blue background that encompassed either the entire cPIC or TFIIH, and these maps were fitting into the reconstructions with a green background that indicate the reconstructions of the entire PIC. Where classes were discarded due to poor quality during classification, the number of particles and the number of classes they encompassed are given as a red bin (number on the bin indicates the number of classes discarded in a given classification run).
Extended Data Fig. 3 Local resolution distribution, FSCs and representative cryo-EM density for the dataset without ADP·BeF3.
a, Reconstructions obtained from the dataset without ADP·BeF3 coloured by their local resolution as estimated using RELION. Total particle count, the global resolution estimate using the Fourier shell correlation (FSC) = 0.143 criterion are given together with an angular distribution plot. b, Solvent-corrected ‘gold-standard’ FSCs grouped according to the reconstructions shown in a. c, Exemplary cryo-EM density (shown as blue mesh) with the fitted atomic models shown with their canonical chain colours. The particular reconstruction and model displayed are specified in the heading of each image. d, Model-to-map FSCs. FSCs (cut-off = 0.5) are grouped according to their resolution-tier going from most focused maps on the top to the overall maps on the bottom.
Extended Data Fig. 4 Upstream flexibility causes variable DNA positions within the cPIC.
a, Structural overlay of the two CC states observed within the PIC dataset without ADP·BeF3 on RPB1. All cPIC components align well except for the upstream complex. Analysis of the difference between these two observed states shows that the upstream complex rotates about the RPB2 protrusion element, and the DNA translates about 7 Å laterally across the cleft. No notable rotation in the DNA was observed, which indicates that this motion is independent of translocation activity. b, Cryo-EM density showing the placement of DNA in the distinct upstream complex classes when the separate classes are aligned on the Pol II core. Red arrows indicate positions of the phosphate backbone in the respective classes. It is notable that in OC-like classes, the motion is of a lesser degree than in the CC classes. The position of TBP is denoted for orientation.
Extended Data Fig. 5 Processing tree of the cryo-EM dataset of PIC with ADP·BeF3.
Processing tree indicating how the particles were assigned their classes and angles for this study. Reconstructions with an orange backing indicate high-resolution maps used for initial building, which were then fitted into reconstructions with a blue background that encompassed either the entire cPIC or TFIIH, and these maps were fitted into the reconstructions with a green background that indicate the reconstructions of the entire PIC. Where classes were discarded due to poor quality during classification, the number of particles and the number of classes they encompassed are given as a red bin (number on the bin indicates the number of classes discarded in a given classification run).
Extended Data Fig. 6 Local resolution distribution, FSCs and representative cryo-EM density for the dataset with ADP·BeF3.
a, Reconstructions obtained from the dataset with ADP·BeF3 coloured by their local resolution as estimated using RELION. Total particle count, the global resolution estimate using the FSC = 0.143 criterion are given together with an angular distribution plot. b, Solvent-corrected ‘gold-standard’ FSCs grouped according to the reconstructions shown in a. c, Exemplary cryo-EM density (shown as blue mesh) with the fitted atomic models shown with their canonical chain colours. The particular reconstruction and model displayed are specified in the heading of each image. d, Model-to-map FSCs. FSCs (cut-off = 0.5) are grouped according to their resolution-tier going from most focused maps on the top to the overall maps on the bottom.
Extended Data Fig. 7 Details of the PIC–TFIIH interaction.
a, Elements of the cPIC and TFIIH involved in maintaining the PIC, and structural comparison of the MAT1 RING finger domain interaction with TFIIE and the Pol II stalk between yeast and human PIC. Although yeast has additional stable contacts such as the PH domain of Tfb1 and E-wing and E-dock, these were not observed for the mammalian PIC. b, Model and cryo-EM density of elements observed for XPB. We observe an N-terminal peptide corresponding to p44 as well as density for a conserved interaction between the TFIIE E-bridge C-terminal anchor.
Extended Data Fig. 8 TFIIH conformational change after ADP·BeF3 binding.
a, Detailed view into the ATP-binding pocket of XPB. Our cryo-EM maps allowed for the unambiguous distinction between the unoccupied pocked (left, without ADP·BeF3) and the ADP·BeF3-bound state with a coordinating magnesium ion. b, Side-by-side comparison of the interactions between XPB and DNA in pre- and post-translocated XPB. Essentially all interactions between XPB and the major or minor grooves are unchanged with the exception of M450 which appears to flip out of the minor groove in the post-translocated state. c, Superposition of the pre- and post-translocated TFIIH on all chains except for XPB, p8 and p52 show the conformational changes in the context of the PIC. Modevectors (right) show that both ATPase lobes of XPB close onto the DNA after ADP·BeF3 binding with lobe 1 moving more than lobe 2. After translocation, the DNA twist stored within XPB is propagated towards cPIC and at the same time a change in trajectory is observed.
Extended Data Fig. 9 Pol II clamp closure and dissociation of the MAT1 RING domain.
a, The closed cPIC viewed from a rotated top view across the closed promoter DNA. The initiator (INR) is found flanked by the Pol II clamp head and the TFIIF charged helix. We observe a partial unwinding of this region after a power-stroke by XPB introduced torsional strain in this area. For clarity, both maps were filtered to a resolution of 5 Å. IC, intermediary complex. b, Conformational change that occurs during the transition between CC and OC. The clamp of Pol II closes upon the DNA, rotating approximately 11° about the Pol II core. Together with the clamp, the stalk of Pol II together with TFIIE buckle inwards. Although a closed clamp is not compatible with closed DNA owing to steric clashes, an open clamp does not clash with open DNA, but is not observed in our data. The CC-to-OC transition in the human PIC also involves restructuring of fork loop 2 from a compact helical conformation to an extended conformation that is not found in the yeast OC (not shown). c, Overview of TFIIB that underwent substantial remodelling. Our model is coloured green, and yeast TFIIB is shown as an outline, and missing secondary structural elements compared with yeast are indicated. Density-to-model fits in four areas of TFIIB (B-ribbon, B-reader, B-linker and B-core), demonstrate excellent agreement of the model to the experimental data. d, Detailed view of the proteinaceous interface between TFIIH and Pol II. The MAT1 RING finger domain nestles between the RPB7 portion of the stalk and on top of the TFIIE ‘E-linker’ helices. Clear cryo-EM density can be observed for the MAT1 RING finger domain in all classes except for the OC class. This indicates that this Pol II–TFIIH interface is disrupted during OC formation but not a direct consequence of XPB translocation, as the intermediary complex class that is in a post-translocated state still shows density for the MAT1 RING finger domain.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1 and Supplementary Figures 1-4.
Video 1
Structure of the mammalian PIC. View of the entire PIC (CC on the left, OC on the right), contact points between cPIC and TFIIH are indicated. For details compare text.
Video 2
Animation of power stroke by XPB translocase. Animation showing morphs between pre- and post-translocated XPB that explains the mechanism of DNA translocation by TFIIH. For details compare text.
Video 3
Animation of promoter opening accompanied by clamp closure. Morph between CC and OC states shows the clamp closure leading to DNA opening as well as MAT1 dissociation. For details compare text.
Rights and permissions
About this article
Cite this article
Aibara, S., Schilbach, S. & Cramer, P. Structures of mammalian RNA polymerase II pre-initiation complexes. Nature 594, 124–128 (2021). https://doi.org/10.1038/s41586-021-03554-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03554-8
This article is cited by
-
The Mediator kinase module enhances polymerase activity to regulate transcriptional memory after heat stress in Arabidopsis
The EMBO Journal (2024)
-
Energy-driven genome regulation by ATP-dependent chromatin remodellers
Nature Reviews Molecular Cell Biology (2024)
-
Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators
Nature Reviews Genetics (2023)
-
Regulation of ribosomal RNA gene copy number, transcription and nucleolus organization in eukaryotes
Nature Reviews Molecular Cell Biology (2023)
-
Structures of transcription preinitiation complex engaged with the +1 nucleosome
Nature Structural & Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.