Abstract
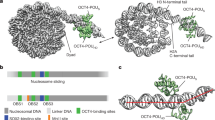
‘Pioneer’ transcription factors are required for stem-cell pluripotency, cell differentiation and cell reprogramming1,2. Pioneer factors can bind nucleosomal DNA to enable gene expression from regions of the genome with closed chromatin. SOX2 is a prominent pioneer factor that is essential for pluripotency and self-renewal of embryonic stem cells3. Here we report cryo-electron microscopy structures of the DNA-binding domains of SOX2 and its close homologue SOX11 bound to nucleosomes. The structures show that SOX factors can bind and locally distort DNA at superhelical location 2. The factors also facilitate detachment of terminal nucleosomal DNA from the histone octamer, which increases DNA accessibility. SOX-factor binding to the nucleosome can also lead to a repositioning of the N-terminal tail of histone H4 that includes residue lysine 16. We speculate that this repositioning is incompatible with higher-order nucleosome stacking, which involves contacts of the H4 tail with a neighbouring nucleosome. Our results indicate that pioneer transcription factors can use binding energy to initiate chromatin opening, and thereby facilitate nucleosome remodelling and subsequent transcription.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM density reconstructions and final models have been deposited with the Electron Microscopy Data Bank (EMD-10390, EMD-10391, EMD-10392, EMD-10393 and EMD-10394) and with the Protein Data Bank (PDB) (6T78, 6T79, 6T7A, 6T7B, 6T7C and 6T7D). All data are available in the Article and its Supplementary Information.
References
Iwafuchi-Doi, M. & Zaret, K. S. Cell fate control by pioneer transcription factors. Development 143, 1833–1837 (2016).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Adachi, K., Suemori, H., Yasuda, S. Y., Nakatsuji, N. & Kawase, E. Role of SOX2 in maintaining pluripotency of human embryonic stem cells. Genes Cells 15, 455–470 (2010).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Fuda, N. J., Ardehali, M. B. & Lis, J. T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461, 186–192 (2009).
Wang, J. et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812 (2012).
Zhu, F. et al. The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81 (2018).
Cirillo, L. A. et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9, 279–289 (2002).
Boller, S., Li, R. & Grosschedl, R. Defining, B cell chromatin: lessons from EBF1. Trends Genet. 34, 257–269 (2018).
Chen, J. et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014).
Velychko, S. et al. Excluding Oct4 from Yamanaka cocktail unleashes the developmental potential of iPSCs. Cell Stem Cell 25, 737–753.e4 (2019).
Soufi, A. et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 (2015).
Meers, M. P., Janssens, D. H. & Henikoff, S. Pioneer factor–nucleosome binding events during differentiation are motif encoded. Mol. Cell 75, 562–575.e5 (2019).
Malik, V. et al. Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat. Commun. 10, 3477 (2019).
Biddle, J. W., Nguyen, M. & Gunawardena, J. Negative reciprocity, not ordered assembly, underlies the interaction of Sox2 and Oct4 on DNA. eLife 8, e41017 (2019).
Kamachi, Y. & Kondoh, H. Sox proteins: regulators of cell fate specification and differentiation. Development 140, 4129–4144 (2013).
Badis, G. et al. Diversity and complexity in DNA recognition by transcription factors. Science 324, 1720–1723 (2009).
Jauch, R., Ng, C. K., Narasimhan, K. & Kolatkar, P. R. The crystal structure of the Sox4 HMG domain–DNA complex suggests a mechanism for positional interdependence in DNA recognition. Biochem. J. 443, 39–47 (2012).
Bergsland, M. et al. Sequentially acting Sox transcription factors in neural lineage development. Genes Dev. 25, 2453–2464 (2011).
Wiebe, M. S., Nowling, T. K. & Rizzino, A. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J. Biol. Chem. 278, 17901–17911 (2003).
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Williams, D. C., Jr, Cai, M. & Clore, G. M. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1–DNA ternary transcription factor complex. J. Biol. Chem. 279, 1449–1457 (2004).
Voong, L. N. et al. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell 167, 1555–1570.e15 (2016).
Li, S., Zheng, E. B., Zhao, L. & Liu, S. Nonreciprocal and conditional cooperativity directs the pioneer activity of pluripotency transcription factors. Cell Rep. 28, 2689–2703 (2019).
Hall, M. A. et al. High-resolution dynamic mapping of histone–DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 16, 124–129 (2009).
Bilokapic, S., Strauss, M. & Halic, M. Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol. 25, 101–108 (2018).
Donovan, B. T., Chen, H., Jipa, C., Bai, L. & Poirier, M. G. Dissociation rate compensation mechanism for budding yeast pioneer transcription factors. eLife 8, e43008 (2019).
Pepenella, S., Murphy, K. J. & Hayes, J. J. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma 123, 3–13 (2014).
Gordon, F., Luger, K. & Hansen, J. C. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 280, 33701–33706 (2005).
Shogren-Knaak, M. et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Dorigo, B., Schalch, T., Bystricky, K. & Richmond, T. J. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 (2003).
Song, F. et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–380 (2014).
Engelen, E. et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 43, 607–611 (2011).
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 (1999).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Li, Z. & Kono, H. Distinct roles of histone H3 and H2A tails in nucleosome stability. Sci. Rep. 6, 31437 (2016).
Lis, J. T. & Schleif, R. Size fractionation of double-stranded DNA by precipitation with polyethylene glycol. Nucleic Acids Res. 2, 383–390 (1975).
Shimko, J. C., North, J. A., Bruns, A. N., Poirier, M. G. & Ottesen, J. J. Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J. Mol. Biol. 408, 187–204 (2011).
Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Bilokapic, S., Strauss, M. & Halic, M. Structural rearrangements of the histone octamer translocate DNA. Nat. Commun. 9, 1330 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 (2007).
Reményi, A. et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17, 2048–2059 (2003).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Klaus, M. et al. Structure and decoy-mediated inhibition of the SOX18/Prox1-DNA interaction. Nucleic Acids Res. 44, 3922–3935 (2016).
Palasingam, P., Jauch, R., Ng, C. K. & Kolatkar, P. R. The structure of Sox17 bound to DNA reveals a conserved bending topology but selective protein interaction platforms. J. Mol. Biol. 388, 619–630 (2009).
Werner, M. H., Huth, J. R., Gronenborn, A. M. & Clore, G. M. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY–DNA complex. Cell 81, 705–714 (1995).
Acknowledgements
We thank members of the Cramer laboratory, in particular H. Hillen, D. Tegunov, and G. Kokic for advice; the crystallization facility at our institute, in particular J. Wawrzinek and U. Steuerwald; and W. Fischle for providing histone expression constructs. Part of this work was performed at Beamline X10SA at the SLS at the PSI. S.O.D. was supported by an EMBO long-term fellowship (ALTF-949-2016). P.C. was supported by the Deutsche Forschungsgemeinschaft (EXC 2067/1-390729940), the ERC Advanced Investigator Grant TRANSREGULON (grant agreement no 693023) and the Volkswagen Foundation.
Author information
Authors and Affiliations
Contributions
S.O.D. designed and carried out all experiments and data analysis. F.Z., supported by J.T., identified the original DNA template used in the study. C.D. assisted with cryo-EM data collection. P.C. designed and supervised research. S.O.D. and P.C. interpreted the data and wrote the manuscript, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 DNA constructs and motif positions.
Related to Fig. 1. a, DNA-1 sequence. Two DNA strands are coloured in dark and light blue, canonical core motifs TTGT are coloured in red, TTXT motifs are shown in orange. Only motifs that allow SOX binding to the DNA minor groove are considered. The position of the third nucleotide of each motif in the DNA-1 sequence is indicated. Motifs at SHL+2 and −2 are shown in bold. b, Top views of the nucleosome. H2A, H2B, H3 and H4 are shown in yellow, red, blue and green, respectively, DNA is shown in dark and light blue. SHLs are labelled. c, DNA-1a template sequence. Only one TTGT motif is present (red). d, Structure of SOX2 (PDB code 1O4X) was aligned to each of the motifs present in nucleosome 1 and allowing binding of SOX to the minor groove. The number of clashes was calculated using ‘findclash’ command in Chimera software. SOX2 binding to motifs at SHL +2 and SHL −2 gives rise to the least amount of clashes with DNA and histones compared to other locations. The ideal position (modelled) of the SOX motif on the dyad would result in a comparably low number of clashes.
Extended Data Fig. 2 Conservation of SOX-family DBD sequence and structure, and X-ray structure of the SOX11–DNA complex.
Related to Fig. 1. a, Domain organization of the human SOX protein family. DBDs are shown as pink rectangles; unstructured functionally diverse regions are shown as wavy lines. Protein constructs used in this study are marked. The alignment of DBD sequences (produced using Clustal Omega) is shown below52. b, Structural conservation of SOX factors. Crystal and nuclear magnetic resonance structures of SOX transcription factors: SOX2 (ref. 22), SOX11 (this study), SOX4 (ref. 18), SOX9, SOX18 (ref. 53) and SOX17 (ref. 54); SRY (ref. 55) has a similar fold. Superimposition of all the structures reveals that they are virtually identical. c, DNA is engulfed by the strongly positively charged inner surface of the SOX11 DBD. d, Ribbon representation of the SOX11 X-ray structure. e, Two copies of SOX11–DNA in the asymmetric unit. The contact between the two is mediated by DNA stacking. f, Comparison of the observed DNA conformation with canonical B-DNA (green). SOX11 introduces a kink into DNA that is typical for HMG box proteins.
Extended Data Fig. 3 EMSAs of SOX2 and SOX11 in complex with DNA or nucleosomes.
Related to Figs. 1–3. EMSAs reveal formation of SOX-factor complexes with DNA (left) or nucleosomes (right). DNA or nucleosome concentration is 1.1 nM. a, EMSA of DNA-1–SOX2 and nucleosome-1–SOX2 complexes. b, EMSA of DNA-1–SOX11 and nucleosome-1–SOX11 complexes. c, EMSA of DNA-1a–SOX11 and nucleosome-1a–SOX11 complexes. d, EMSA of DNA–Widom+1 and nucleosome–Widom+1–SOX11 complexes. e, EMSA of DNA Widom 601–SOX11 and nucleosome Widom 601–SOX11 complexes. Relevant bands are labelled. All experiments were repeated at least twice with similar results. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4 Global and local resolution of cryo-EM reconstructions.
Related to Figs. 1–3. a, Example micrograph reveals preferred orientation of nucleosomes. Scale bar, 20 nm. b, FSC plots for five reported reconstructions. c, Local resolution distribution. In some maps, the resolution varies from 3 Å (dark blue) through 4 Å (green) to 6 Å (red). The lower resolution of some DNA regions indicates flexibility. d, Angular distribution plots. Scale shows the number of particles assigned to a particular angle. Blue, a low number of particles; yellow, a high number of particles. e, Directional FSC plots for the reconstructions calculated on the 3DFSC server48. Sphericity, as the degree of anisotropy present in the reconstructions, is indicated above each plot. Histograms indicate the portion of voxels with a particular resolution. f, Visualization of different regions of the nucleosome–SOX11 map. In the nucleosome core, histone side chains are clearly visible; SOX density has a lower resolution, but helical densities are clearly distinguishable. g, Rigid-body fit of the SOX2–DNA structure (PDB code 1O4X) into the nucleosome–SOX2 cryo-EM map (left). Rigid-body fit of the SOX11–DNA X-ray structure into the nucleosome–SOX11 cryo-EM map (right). The region containing SOX and a short DNA stretch was isolated from the rest of the map for clarity.
Extended Data Fig. 6 EMSAs using SOX2, and wild-type or mutant SOX11 in complex with DNA or nucleosomes.
Related to Figs. 1–3. EMSAs reveal the formation of SOX-factor complexes with DNA or nucleosomes. All experiments were repeated at least twice with similar results. For gel source data, see Supplementary Fig. 1. a, EMSA of DNA-1 or nucleosome-1 with wild-type SOX2. b, EMSA of DNA-1 or nucleosome-1 with wild-type SOX11. c, EMSA of DNA-1 or nucleosome-1 with SOX11(H75A/S80A). d, EMSA of DNA-1 or nucleosome-1 with SOX11(Y118A). Relevant bands are labelled. To observe a wider range of the binding curve for the mutants, a larger amount of glycerol (12% final concentration) was used here (as compared to the EMSAs shown in Extended Data Fig. 3, which used 5% glycerol)—thus, the apparent affinity is higher. DNA or nucleosome concentration is 1.1 nM. e, EMSAs reveal formation of SOX–DNA or SOX–nucleosome complexes at different concentrations of salt. There is virtually no difference in binding at 10 or 30 mM NaCl, whereas binding is weaker at 150 mM NaCl. DNA or nucleosome concentration is 1.1 nM. f, EMSAs reveal formation of SOX complexes with DNA-1a or nucleosome-1a (only one canonical motif present) as compared to DNA-1b and nucleosome-1b (in which all canonical SOX motifs were mutated). SOX concentrations ranged from 0 to 200 nM, DNA concentration was 1.1 nM. g, Superimposition of two nucleosome–SOX11 cryo-EM maps obtained in the presence of 1 mM MgCl2 (grey density) or in the absence of MgCl2 (coloured). Magnesium does not influence the structure of the SOX–nucleosome complex. Cross-correlation between the two unmasked maps is 0.94 (Chimera).
Extended Data Fig. 7 Nucleosome-DNA end unwrapping.
Related to Figs. 1, 3. a, DNA-1 sequence with BfuCI digestion sites. BfuCI digestion assays (two independent experiments for each sample, shown here) of the DNA-1, nucleosome-1 or nucleosome-1b (no TTGT motifs) in the presence of increasing amounts of SOX11. The restriction site (about 20 nucleotides away from the entry and exit sites of the nucleosome) becomes more accessible with higher concentrations of SOX11. In a DNA-1 digestion assay, the amount of digestion product increases slightly in the low SOX11 concentration range, and then stays constant over a broad concentration range. For gel source data, see Supplementary Fig. 1. b, BfuCI digestion assay plot for free DNA-1 (black), nucleosome-1 (green) and mutated nucleosome-1b (orange) in the presence of increasing amounts of SOX11. Each experiment was performed independently twice (n = 2). Mean values (lines) and individual measurements (dots) are shown. Band intensity was calculated using standard routine in ImageJ52. c, Example fluorescence spectra of Cy3–Cy5-labelled nucleosome in the presence of increasing amounts of SOX11. d, FRET efficiency plot. Mean values with s.d. are shown (independent experiments, n = 4).
Extended Data Fig. 8 Repositioning of the H4 N-terminal tail.
Related to Fig. 4. Cryo-EM maps are shown with a Gaussian smoothening filter (Chimera49) applied for clarity. SOX is coloured in pink, H4 is shown in green and the repositioned H4 tail is shown in orange. a, Free nucleosome. Views of SHL +2 and SHL −2 are shown to illustrate the position of the H4 tail (a dashed line). Residue K16 is marked with a circle. b, Nucleosome–SOX11 with SOX11 located at SHL +2. On the right, a superimposition with the free nucleosome map is shown to highlight different orientations of the H4 tail. c, Nucleosome–SOX11 complex with SOX11 located at SHL −2. In this location, SOX is oriented differently and does not clash with or reposition the H4 tail. d, Nucleosome–SOX11*. The H4 N-terminal tail is repositioned compared with that in the free nucleosome. Repositioning of the H4 tail was reported in case of strong distortions in the nucleosome structure30. e, SOX binding repositions the H4 N-terminal tail and might impair nucleosome stacking. Side view of two stacking nucleosomes (from PDB code 1AOI). H4 interacts with the acidic patch on the neighbouring H2A–H2B histone dimer. H4 tail repositioning is incompatible with nucleosome stacking.
Supplementary information
Supplementary Figure 1
Raw images of gels related to Extended Data Figs 3, 6 and 7. The cropping borders are indicated with dashed lines.
Video 1: Overview of Nucleosome-Sox complexes
Pioneer factor binding remodels the nucleosome. Binding of Sox11 to the nucleosome leads to local DNA distortion and unwrapping of terminal DNA. The five cryo-EM structures reported here have been used to generate this animation. For details compare text.
Rights and permissions
About this article
Cite this article
Dodonova, S.O., Zhu, F., Dienemann, C. et al. Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function. Nature 580, 669–672 (2020). https://doi.org/10.1038/s41586-020-2195-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2195-y
This article is cited by
-
Structural basis of Integrator-dependent RNA polymerase II termination
Nature (2024)
-
Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBPα
Nature Structural & Molecular Biology (2024)
-
DNA binding redistributes activation domain ensemble and accessibility in pioneer factor Sox2
Nature Communications (2024)
-
Pioneer factor Pax7 initiates two-step cell-cycle-dependent chromatin opening
Nature Structural & Molecular Biology (2024)
-
Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition
Nature Structural & Molecular Biology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.