Abstract
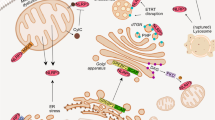
The cellular stress response has a vital role in regulating homeostasis by modulating cell survival and death. Stress granules are cytoplasmic compartments that enable cells to survive various stressors. Defects in the assembly and disassembly of stress granules are linked to neurodegenerative diseases, aberrant antiviral responses and cancer1,2,3,4,5. Inflammasomes are multi-protein heteromeric complexes that sense molecular patterns that are associated with damage or intracellular pathogens, and assemble into cytosolic compartments known as ASC specks to facilitate the activation of caspase-1. Activation of inflammasomes induces the secretion of interleukin (IL)-1β and IL-18 and drives cell fate towards pyroptosis—a form of programmed inflammatory cell death that has major roles in health and disease6,7,8,9,10,11,12. Although both stress granules and inflammasomes can be triggered by the sensing of cellular stress, they drive contrasting cell-fate decisions. The crosstalk between stress granules and inflammasomes and how this informs cell fate has not been well-studied. Here we show that the induction of stress granules specifically inhibits NLRP3 inflammasome activation, ASC speck formation and pyroptosis. The stress granule protein DDX3X interacts with NLRP3 to drive inflammasome activation. Assembly of stress granules leads to the sequestration of DDX3X, and thereby the inhibition of NLRP3 inflammasome activation. Stress granules and the NLRP3 inflammasome compete for DDX3X molecules to coordinate the activation of innate responses and subsequent cell-fate decisions under stress conditions. Induction of stress granules or loss of DDX3X in the myeloid compartment leads to a decrease in the production of inflammasome-dependent cytokines in vivo. Our findings suggest that macrophages use the availability of DDX3X to interpret stress signals and choose between pro-survival stress granules and pyroptotic ASC specks. Together, our data demonstrate the role of DDX3X in driving NLRP3 inflammasome and stress granule assembly, and suggest a rheostat-like mechanistic paradigm for regulating live-or-die cell-fate decisions under stress conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The proteomics data generated and analysed during the current study are available in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014828. Raw data for figures are available as Source Data to the relevant figure. All other datasets generated and analysed during the current study are available from the corresponding authors on reasonable request. Any requests for data or materials should, in the first instance, be addressed to T.-D.K.
References
Protter, D. S. W. & Parker, R. Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016).
Wolozin, B. Regulated protein aggregation: stress granules and neurodegeneration. Mol. Neurodegener. 7, 56 (2012).
White, J. P. & Lloyd, R. E. Regulation of stress granules in virus systems. Trends Microbiol. 20, 175–183 (2012).
Anderson, P. & Kedersha, N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10, 430–436 (2009).
Anderson, P., Kedersha, N. & Ivanov, P. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta 1849, 861–870 (2015).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Kanneganti, T.-D. et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440, 233–236 (2006).
Mariathasan, S. et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 (2006).
Karki, R. & Kanneganti, T.-D. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 19, 197–214 (2019).
Malireddi, R. K. S. et al. TAK1 restricts spontaneous NLRP3 activation and cell death to control myeloid proliferation. J. Exp. Med. 215, 1023–1034 (2018).
Man, S. M. & Kanneganti, T.-D. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16, 7–21 (2016).
Venegas, C. et al. Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature 552, 355–361 (2017).
Muñoz-Planillo, R. et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013).
Kanneganti, T.-D. & Lamkanfi, M. K+ drops tilt the NLRP3 inflammasome. Immunity 38, 1085–1088 (2013).
Groß, C. J. et al. K+ efflux-independent NLRP3 inflammasome activation by small molecules targeting mitochondria. Immunity 45, 761–773 (2016).
Lu, A. et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 (2014).
Franklin, B. S. et al. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat. Immunol. 15, 727–737 (2014).
Franklin, B. S., Latz, E. & Schmidt, F. I. The intra- and extracellular functions of ASC specks. Immunol. Rev. 281, 74–87 (2018).
Kedersha, N. et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 (2005).
Man, S. M. et al. IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell 167, 382–396 (2016).
Bol, G. M. et al. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol. Med. 7, 648–669 (2015).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Aditi, F., Folkmann, A. W. & Wente, S. R. Cytoplasmic hGle1A regulates stress granules by modulation of translation. Mol. Biol. Cell 26, 1476–1490 (2015).
Hilliker, A., Gao, Z., Jankowsky, E. & Parker, R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 43, 962–972 (2011).
Shih, J.-W. et al. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 441, 119–129 (2012).
Cruciat, C.-M. et al. RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-β-catenin signaling. Science 339, 1436–1441 (2013)
Soulat, D. et al. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 27, 2135–2146 (2008).
Stunnenberg, M., Geijtenbeek, T. B. H. & Gringhuis, S. I. DDX3 in HIV-1 infection and sensing: a paradox. Cytokine Growth Factor Rev. 40, 32–39 (2018).
Robinson, G. et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48 (2012).
Ditton, H. J., Zimmer, J., Kamp, C., Rajpert-De Meyts, E. & Vogt, P. H. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum. Mol. Genet. 13, 2333–2341 (2004).
Vakilian, H. et al. DDX3Y, a male-specific region of Y chromosome gene, may modulate neuronal differentiation. J. Proteome Res. 14, 3474–3483 (2015).
Chen, C.-Y. et al. Targeted inactivation of murine Ddx3x: essential roles of Ddx3x in placentation and embryogenesis. Hum. Mol. Genet. 25, 2905–2922 (2016).
Li, Q. et al. DDX3X regulates cell survival and cell cycle during mouse early embryonic development. J. Biomed. Res. 28, 282–291 (2014).
Jones, J. W. et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl Acad. Sci. USA 107, 9771–9776 (2010).
Franchi, L. et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat. Immunol. 7, 576–582 (2006).
Van Gorp, H. et al. Familial Mediterranean fever mutations lift the obligatory requirement for microtubules in Pyrin inflammasome activation. Proc. Natl Acad. Sci. USA 113, 14384–14389 (2016).
Wheeler, J. R., Matheny, T., Jain, S., Abrisch, R. & Parker, R. Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 (2016).
Szaflarski, W. et al. Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget 7, 30307–30322 (2016).
Karki, R. et al. IRF8 regulates transcription of Naips for NLRC4 inflammasome activation. Cell 173, 920–933 (2018).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protocols 11, 2301–2319 (2016).
Kesavardhana, S. et al. ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. 214, 2217–2229 (2017).
Buchan, D. W. A. & Jones, D. T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 47 (W1), W402–W407 (2019).
Acknowledgements
We thank all the members of the Kanneganti laboratory for their comments and suggestions; F. Phillips and N. Lantz for their help with generating BMDMs and with mouse husbandry; K. A. Laycock and R. Tweedell for scientific editing of the manuscript; K. Kodali and V. Pagala for their help with affinity purification mass spectrometry analysis; and S. Miller and S. Sakurada for helping with the CRISPR–Cas9-mediated gene disruption experiment. psPAX2 (Addgene plasmid 12260) and pMD2.G (Addgene plasmid 1225) were gifts from D. Trono. T.-D.K. is supported by NIH grants AI101935, AI124346, AR056296 and CA163507 and by the American Lebanese Syrian Associated Charities; the St. Jude Children's Research Hospital Cell and Tissue Imaging Center is supported by St. Jude Children's Research Hospital and by National Cancer Institute grant P30 CA021765-35; R.J.G. is supported by Cancer Research UK, the Mathile Family Foundation, Cure Search, the Sohn Foundation and NIH grants P01CA96832 and R0CA1129541.
Author information
Authors and Affiliations
Contributions
P.S., S.K. and T.-D.K. conceptualized the study and designed the experiments; D.M.P., S.G., S.P. and R.J.G. generated and characterized the Ddx3xfl/fl mice; P.S., S.K., D.M.P., R.K.S.M., R.K., C.S.G., B.B., D.E.P., A.B., B.R.S., A.N., S.V.K., A.P. and A.R.B. performed the experiments; P.S., S.K., R.K.S.M., R.K. and T.-D.K. conducted the analyses; P.S., S.K. and T.-D.K. wrote the manuscript; and T.-D.K. supervised the project and provided guidance.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
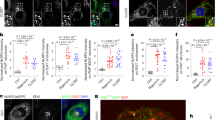
Extended Data Fig. 1 Specific inhibition of the NLRP3 inflammasome is not due to differences in levels of core inflammasome component proteins, NLRP3 sequestration in stress granules or induction of autophagy.
a, Confocal microscopy imaging of LPS-primed BMDMs treated with arsenite (50 μM) or with anisomycin (25 μg ml−1) and arsenite. Scale bars, 10 μm. Representative images (n = 2). b, Quantification of ASC+ cells stimulated with LPS and treated with nigericin, arsenite and anisomycin as indicated. ***P = 0.0002 (unpaired two-sided t-test). Data are mean ± s.e.m. (n > 70 frames). c, Immunoblot analysis of pro-CASP1 (p45) and cleaved CASP1 (p20) in BMDMs treated with poly(I:C) (P(I:C)); poly(I:C) and arsenite; poly(I:C) and nigericin; or poly(I:C), arsenite and nigericin. Representative blots (n = 2 biologically independent experiments). d, Quantification of inflammatory cytokines in BMDMs treated with medium (Mock), LPS, LPS and arsenite, or arsenite. P values for LPS versus LPS + Ar: IL-6, P = 0.8561; KC, P = 0.9522; TNF, P = 0.2599. Data are mean ± s.e.m. (n = 4). e, f, Immunoblot analysis of CASP1 cleavage in BMDMs treated with the AIM2-activating ligand poly(dA:dT) (pA:T) (e) or the NLRC4-activating ligand flagellin (Fla) (f). Representative blots (n = 3 biologically independent experiments each). g, Immunoblot analysis of protein levels of core NLRP3 inflammasome components after LPS priming, arsenite-mediated induction of stress granules and anisomycin-mediated inhibition of stress granules. Representative blots (n = 3 biologically independent experiments). h, Confocal microscopy imaging of BMDMs treated with LPS alone or with LPS and arsenite to visualize NLRP3 localization. G3BP1 was used as a marker of stress granules. Scale bars, 10 μm (whole-cell images); 5 μm (magnified images). Representative images (n = 2 biologically independent experiments). i, Immunoblot analysis of CASP1 cleavage in autophagy-deficient BMDMs that lack Becn1 and Atg5, stimulated with LPS and treated with nigericin, arsenite and anisomycin as indicated. Representative blots (n = 2 biologically independent experiments).
Extended Data Fig. 2 DDX3X colocalizes with G3BP1 in stress granules in BMDMs treated with arsenite.
a, Volcano plot of the affinity purification mass spectrometry analysis of the NLRP3 interactome, with NLRP3 (green) and DDX3X (red) highlighted. Two biologically independent replicates each of samples treated with LPS and samples treated with LPS and nigericin were pooled together for analysis. Data can be accessed using ProteomeXchange with the identifier PXD014828. b, c, Structured illumination microscopy of LPS-primed BMDMs treated with or without arsenite to visualize DDX3X, the stress granule marker G3BP1 (b) and ASC (c). Scale bars, 3 μm. Representative images (n = 2 each).
Extended Data Fig. 3 Strategy for generating the Ddx3x gene knockout and characterization of immune cells.
a, Genomic locations of loxP sites in the Ddx3x locus. b, Genotyping PCR for the 5′ and 3′ loxP sites. Representative gel (n = 1). L, molecular-weight ladder. c, Immunoblot analysis of the levels of DDX3X protein in BMDMs. Data are from three different mice with each genotype. d, Flow cytometry analysis of ex vivo differentiated BMDMs stained for CD11b and F4/80. e, Quantification of the numbers of different immune-cell types from the blood of the indicated mouse strains. RBC, red blood cells. Data are mean ± s.e.m. (n = 3).
Extended Data Fig. 4 Lack of DDX3X leads to defects in both stress granule assembly and NLRP3 inflammasome activation.
a, Confocal microscopy of Ddx3xfl/fl and Ddx3xfl/flLysMcre BMDMs stimulated with LPS and arsenite to visualize localization of the stress granule marker G3BP1 and DDX3X. Scale bars, 10 μm. b, Quantification of the number of stress granules per cell. Each data point represents the average number of stress granules per cell in each field of view. ***P = 0.0003 (unpaired two-sided t-test). Data are mean ± s.e.m. (n = 10). c, Immunoblot analysis of CASP1 cleavage in peritoneal macrophages stimulated with LPS with or without nigericin. Representative blots (n = 2 biologically independent experiments). d, Pyroptotic cell death as measured by the number of SYTOX Green+ cells. Cells were stimulated with LPS and treated with nigericin or with nigericin and arsenite. ****P ≤ 0.0001 (two-way ANOVA) for LPS + N-treated Ddx3xfl/fl versus LPS + N-treated Ddx3xfl/flLysMcre BMDMs; LPS + N-treated versus LPS + Ar + N-treated Ddx3xfl/fl BMDMs; and LPS + N-treated versus LPS + Ar + N-treated Ddx3xfl/flLysMcre BMDMs. Data are mean ± s.e.m. (n = 4). e–g, Immunoblot analysis of CASP1 cleavage in BMDMs treated with LPS or LPS and ATP (e), Pam3CSK4 or Pam3CSK4 and nigericin (f), or poly(I:C) or poly(I:C) and nigericin (g). Representative blots (n = 2 biologically independent experiments each).
Extended Data Fig. 5 Loss of DDX3X in BMDMs leads to the specific inhibition of NLRP3 inflammasome activation.
a–d, CASP1 cleavage in stimulated and unstimulated BMDMs and IL-1β and IL-18 release from BMDMs stimulated to activate the NLRP3, NLRC4, AIM2 and PYRIN inflammasomes by using LPS and nigericin (a) (ELISA, n > 10); flagellin (b) (ELISA, n = 2); poly(dA:dT) (c) (ELISA, n = 2); and Clostridium difficile toxin B (TcdB) (d) (ELISA, n = 2). P values in a (from left to right): *P = 0.0170, *P = 0.0116 (unpaired two-sided t-test); n.d., not detected. ELISA data are mean ± s.e.m. Representative blots (n = 3 biologically independent experiments each). e, Immunoblot analysis of CASP1 cleavage in wild-type BMDMs expressing a doxycycline-inducible DDX3X–mCherry construct and treated with LPS and nigericin. Representative blots (n = 2 biologically independent experiments). f, Immunoblot analysis of CASP1 cleavage in wild-type BMDMs and Ddx3xfl/flLysMcre BMDMs that constitutively express DDX3X–YFP from a cytomegalovirus promoter, treated with LPS and nigericin. Representative blots (n = 2 biologically independent experiments).
Extended Data Fig. 6 DDX3X interacts with NLRP3 but not ASC and CASP1, and its loss does not lead to a decrease in the levels of core components of the NLRP3 inflammasome.
a, Immunoblot analysis of the levels of NLRP3, ASC, CASP1, pro-IL-1β, NEK7, DDX3X and GAPDH proteins in Ddx3xfl/fl, Ddx3xfl/flLysMcre and Nlrp3−/− BMDMs after LPS priming. Representative blots (n = 3 biologically independent experiments). b, Immunoblot analysis of NLRP3 and DDX3X immunoprecipitation from HEK293T cells transfected with Flag–NLRP3 and DDX3X–mCherry expression constructs. S.E., short exposure; L.E., long exposure. Representative blots (n = 3 biologically independent experiments). c, Immunoblot analysis of ASC and DDX3X immunoprecipitates from HEK293T cells transfected with ASC and DDX3X–mCherry expression constructs. Representative blots (n = 3 biologically independent experiments). d, Immunoblot analysis of CASP1 and DDX3X immunoprecipitates from HEK293T cells transfected with CASP1 and DDX3X–mCherry expression constructs. Representative blots (n = 2 biologically independent experiments). e, Quantification of the percentage of cells that contain ASC specks in Ddx3xfl/fl and Ddx3xfl/flLysMcre BMDMs. ****P ≤ 0.0001 (unpaired two-sided t-test). Data are mean ± s.e.m. (n = 48). f, Quantification of the percentage of cells that contain ASC specks in wild-type and Ddx3xfl/flLysMcre peritoneal macrophages (PMs). ****P ≤ 0.0001 (unpaired two-sided t-test). Data are mean ± s.e.m. (n = 48). g, Confocal microscopy imaging of wild-type and Ddx3xfl/flLysMcre peritoneal macrophages treated with LPS and nigericin to visualize ASC and DDX3X. Scale bar, 10 μm.
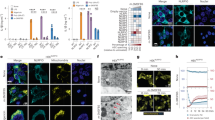
Extended Data Fig. 7 Induction of stress granules by various stressors.
a–d, Immunoblot analysis of CASP1 cleavage in LPS-primed BMDMs. Representative blots (n = 3 biologically independent experiments each). a, BMDMs were treated with 2 μg ml−1 thapsigargin (Th) for 1 h before NLRP3 inflammasome activation by nigericin. b, BMDMs were treated with 400 μM d-sorbitol (Sb) for 30 min before NLRP3 inflammasome activation by nigericin. c, BMDMs were treated with 400 μM paclitaxel (PCX) for 30 min before NLRP3 inflammasome activation by nigericin. d, BMDMs were treated with 750 μM vincristine (VCR) for 30 min before NLRP3 inflammasome activation by nigericin. e, f, Confocal microscopy images to visualize the induction of stress granules in BMDMs (e) and L929 cells (f) using 50 μM arsenite, 2 μg ml−1 thapsigargin, 400 μM d-sorbitol, 400 μM paclitaxel, 750 μM vincristine and 5 μM RK-33. Cells were incubated with stressors for 1 h and stained with the stress granule marker G3BP1 and with DDX3X. Representative images (n = 3 biologically independent experiments each).
Extended Data Fig. 8 Identification of the region of DDX3X required for its interaction with NLRP3, prediction of disordered regions using PONDR and PSIPRED, and effect of DDX3X helicase activity inhibition on NLRP3 inflammasome activation.
a, Schematic of C-terminal mCherry-tagged DDX3X domain-deletion expression constructs. b, Immunoblot analysis of the input lysates used for immunoprecipitation of Flag–NLRP3-FL and DDX3X–mCherry constructs. Representative blots (n = 2 biologically independent experiments). c–f, Immunoblot analysis of immunoprecipitates with Flag–NLRP3-FL and mCherry-tagged DDX3X domain-deletion expression constructs: full-length DDX3X (c), DDX3X with an N-terminal deletion (d), DDX3X with a C-terminal deletion (e) and the DDX3X helicase domain (f). Red asterisk indicates a non-specific band. Representative blots (n = 2 biologically independent experiments each). g, Overlay of the predicted disordered regions in NLRP3 from PONDR and PSIPRED. h, Overlay of the predicted disordered regions in DDX3X from PONDR and PSIPRED. i, Immunoblot analysis of CASP1 cleavage in BMDMs treated with LPS; LPS and nigericin; LPS and RK-33; LPS, RK-33 and arsenite; LPS, arsenite and nigericin; LPS, RK-33 and nigericin; and LPS, RK-33, arsenite and nigericin. Representative blots (n = 2 biologically independent experiments).
Extended Data Fig. 9 Stress granule-mediated inhibition of oligomerization of NLRP3 and ASC and induction of NLRP3 aggregation.
a, Immunoblot analysis of crosslinked and un-crosslinked (BME-treated) lysates of HEK293T cells expressing mouse ASC, DDX3X and NLRP3. Representative blots (n = 3). WCE, whole-cell extracts. b, Immunoblot analysis of crosslinked and non-reduced (that is, without BME treatment) BMDM lysates. Representative blots (n = 3). c, d, Secretion of inflammasome-dependent cytokines IL-1β (c) and IL-18 (d) from BMDMs treated with arsenite for different durations before the addition of nigericin to activate the NLRP3 inflammasome. Arsenite was added 30 or 15 min before, simultaneously with or 15 or 30 min after nigericin. Samples were collected after 75 min of nigericin treatment. For the anisomycin-treated samples, anisomycin was given 20 min before the start of the earliest arsenite treatment (−30 min). A sigmoid curve was fitted to the data to show the correlation between the timing of arsenite addition and the release of inflammasome-dependent cytokines. The goodness of fit was determined by the R2 value. P values (for the column factor that represents anisomycin treatment): **P = 0.0010 (c) and **P = 0.0066 (d) by two-way ANOVA. Data are mean ± s.e.m. (n = 3).
Extended Data Fig. 10 Effects of the loss of G3BP1 on NLRP3 inflammasome activation and intraperitoneal injection of arsenite on the release of inflammatory cytokines.
a, Immunoblot analysis of CASP1 cleavage, total G3BP1 and GAPDH in wild-type and dCas9-expressing immortalized BMDMs transduced with two different gRNAs that target G3bp1. Representative blots (n = 3). b, c, Quantification of inflammatory cytokines in an LPS-induced peritonitis model with or without intraperitoneal injection of arsenite, in serum (b) and in peritoneal fluid (c). P values: **P = 0.0051 (IP-10 in serum), **P = 0.0024 (RANTES in peritoneal fluid) by unpaired two-sided t-test. Data are mean ± s.e.m. (n ≥ 6).
Supplementary information
Supplementary Figure
Uncropped western blot images with molecular weight markers and indication of how the gels were cropped.
Source data
Rights and permissions
About this article
Cite this article
Samir, P., Kesavardhana, S., Patmore, D.M. et al. DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594 (2019). https://doi.org/10.1038/s41586-019-1551-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1551-2
This article is cited by
-
Multiomics characterization of pyroptosis in the tumor microenvironment and therapeutic relevance in metastatic melanoma
BMC Medicine (2024)
-
DDX3X interacts with SIRT7 to promote PD-L1 expression to facilitate PDAC progression
Oncogenesis (2024)
-
miR-181a-5p targets DDX3X to inhibit the progression of osteoarthritis via NF-ΚB signaling pathway
Journal of Orthopaedic Surgery and Research (2023)
-
Stress granule activation attenuates lipopolysaccharide-induced cardiomyocyte dysfunction
BMC Cardiovascular Disorders (2023)
-
NLRP3 inflammasome in cognitive impairment and pharmacological properties of its inhibitors
Translational Neurodegeneration (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.