Abstract
A decline in stem cell function impairs tissue regeneration during ageing, but the role of the stem-cell-supporting niche in ageing is not well understood. The small intestine is maintained by actively cycling intestinal stem cells that are regulated by the Paneth cell niche1,2. Here we show that the regenerative potential of human and mouse intestinal epithelium diminishes with age owing to defects in both stem cells and their niche. The functional decline was caused by a decrease in stemness-maintaining Wnt signalling due to production of Notum, an extracellular Wnt inhibitor, in aged Paneth cells. Mechanistically, high activity of mammalian target of rapamycin complex 1 (mTORC1) in aged Paneth cells inhibits activity of peroxisome proliferator activated receptor α (PPAR-α)3, and lowered PPAR-α activity increased Notum expression. Genetic targeting of Notum or Wnt supplementation restored function of aged intestinal organoids. Moreover, pharmacological inhibition of Notum in mice enhanced the regenerative capacity of aged stem cells and promoted recovery from chemotherapy-induced damage. Our results reveal a role of the stem cell niche in ageing and demonstrate that targeting of Notum can promote regeneration of aged tissues.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
RNA sequencing data are publicly available through ArrayExpress with accession code E-MTAB-7916. Source Data for Fig. 1–3 and Extended Data Figs. 1–9 are available with the online version of the paper. All other data are available from the corresponding author upon reasonable request.
References
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Sato, T. et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011).
Sengupta, S., Peterson, T. R., Laplante, M., Oh, S. & Sabatini, D. M. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104 (2010).
Molofsky, A. V. et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443, 448–452 (2016).
Rossi, D. J. et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 (2007).
Conboy, I. M. & Rando, T. A. Heterochronic parabiosis for the study of the effects of aging on stem cells and their niches. Cell Cycle 11, 2260–2267 (2012).
Sato, T. et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Warren, P. M., Pepperman, M. A. & Montgomery, R. D. Age changes in small-intestinal mucosa. Lancet 312, 849–850 (1978).
Feibusch, J. M. & Holt, P. R. Impaired absorptive capacity for carbohydrate in the aging human. Dig. Dis. Sci. 27, 1095–1100 (1982).
Feldman, M., Cryer, B., McArthur, K. E., Huet, B. A. & Lee, E. Effects of aging and gastritis on gastric acid and pepsin secretion in humans: a prospective study. Gastroenterology 110, 1043–1052 (1996).
Potten, C. S., Martin, K. & Kirkwood, T. B. Ageing of murine small intestinal stem cells. Novartis Found Symp. 235, 66–79 (2001).
Yilmaz, O. H. et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012).
Nalapareddy, K. et al. Canonical Wnt signaling ameliorates aging of intestinal stem cells. Cell Rep. 18, 2608–2621 (2017).
Mihaylova, M. M. et al. Fasting activates fatty acid oxidation to enhance intestinal stem cell function during homeostasis and aging. Cell Stem Cell 22, 769–778 (2018).
Giráldez, A. J., Copley, R. R. & Cohen, S. M. HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev. Cell 2, 667–676 (2002).
Kakugawa, S. et al. Notum deacylates Wnt proteins to suppress signalling activity. Nature 519, 187–192 (2015).
Shoshkes-Carmel, M. et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246 (2018).
Degirmenci, B., Valenta, T., Dimitrieva, S., Hausmann, G. & Basler, K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 558, 449–453 (2018).
Farin, H. F., Van Es, J. H. & Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529 (2012).
Farin, H. F. et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340–343 (2016).
McCay, C. M., Maynard, L. A., Sperling, G. & Barnes, L. L. Retarded growth, life span, ultimate body size and age changes in the albino rat after feeding diets restricted in calories. Nutr. Rev. 33, 241–243 (1975).
Harrison, D. E. et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 (2009).
Lamming, D. W. et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643 (2012).
Naillat, F. et al. Identification of the genes regulated by Wnt-4, a critical signal for commitment of the ovary. Exp. Cell Res. 332, 163–178 (2015).
Suciu, R. M., Cognetta, A. B., III, Potter, Z. E. & Cravatt, B. F. Selective irreversible inhibitors of the Wnt-deacylating enzyme NOTUM developed by activity-based protein profiling. ACS Med. Chem. Lett. 9, 563–568 (2018).
Longley, D. B., Harkin, D. P. & Johnston, P. G. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Cancer 3, 330–338 (2003).
Song, M. K., Park, M. Y. & Sung, M. K. 5-fluorouracil-induced changes of intestinal integrity biomarkers in BALB/c mice. J. Cancer Prev. 18, 322–329 (2013).
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Kim, T. H., Escudero, S. & Shivdasani, R. A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl Acad. Sci. USA 109, 3932–3937 (2012).
Zou, W. Y. et al. Epithelial WNT ligands are essential drivers of intestinal stem cell activation. Cell Rep. 22, 1003–1015 (2018).
Kozar, S. et al. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell 13, 626–633 (2013).
Frey, J. L. et al. Wnt–Lrp5 signaling regulates fatty acid metabolism in the osteoblast. Mol. Cell. Biol. 35, 1979–1991 (2015).
Huels, D. J. et al. Wnt ligands influence tumour initiation by controlling the number of intestinal stem cells. Nat. Commun. 9, 1132 (2018).
Beyaz, S. et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58 (2016).
Chang, S., Goldstein, N. E. & Dharmarajan, K. V. Managing an older adult with cancer: considerations for radiation oncologists. BioMed Res. Int. 2017, 1695101 (2017).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
Chen, B. et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 (2009).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015).
Dahlman, J. E. et al. Orthogonal gene knockout and activation with a catalytically active Cas9 nuclease. Nat. Biotechnol. 33, 1159–1161 (2015).
Sánchez-Rivera, F. J. et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 17.1.2001 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9, e1003118 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Adhikary, T. et al. Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-β/δ (PPARβ/δ). PLoS ONE 6, e16344 (2011).
Kwiatkowski, D. J. et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11, 525–534 (2002).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Roper, J. et al. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat. Protoc. 13, 217–234 (2018).
Roper, J. et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017).
Acknowledgements
This study was supported by the Academy of Finland (Research Fellow and Centre of Excellence, MetaStem), Marie Curie CIG (618774), ERC-STG (677809), Swedish Research Council 2018-03078, Sigrid Juselius Foundation, Center for Innovative Medicine, and Wallenberg Academy Fellows program to P.K. N.P. was supported by the Integrative Life Science Doctoral program and by the Research foundation of University of Helsinki. B.F.C. was supported by grant R35 CA231991. We thank the personnel of the DNA sequencing and genomics laboratory for performing the RNA sequencing assays. We thank J. Bärlund, A. Sola-Carvajal, M. Simula and A. Kegel for technical assistance.
Author information
Authors and Affiliations
Contributions
N.P. and P.K. designed and interpreted the results of all experiments. N.P., S.I., M.M., S.A., A.B.C. III, R.M.S., K.L., E.M., S.G., S.N. and T.T. performed all experiments and analysed the results. J.R. performed and analysed organoid transplantations. O.-P.S. processed and analysed the RNA sequencing data with the help of P.A., P.K. and N.P. N.P., M.M., K.L., E.M., S.A. and S.G. performed and analysed the immunohistochemistry, immunofluorescence and RNA in situ hybridizations. N.G., T.S., A.J., K.P. and A.R. provided the human biopsy material. Ö.H.Y., D.M.S., T.T., T.J. and B.F.C. participated in the design and interpretation of experiments. P.K. and N.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Characterization of aged intestine.
a, Organoid-forming capacity of crypts from young and old mice (n = 4 mice per group). Student’s paired t-test. b, Frequency of organoids unable to form new crypts (fission deficiency) in young and old mice (n = 6 mice per group) analysed 5–9 days after isolation. Student’s paired t-test. c, Distribution of regenerative growth capacity of primary organoids from young and old mice (n = 6 mice per group). d, Regenerative growth of subcultured secondary mouse organoids (n = 6 mice per group). Student’s paired t-test. e, Distribution of regenerative growth capacity of subcultured secondary organoids from old and young mice (n = 6 mice per group). f, Representative H&E staining of mouse jejunal sections from young and old mice (four mice analysed per group). g, Quantification of EdU+ cells in jejunal crypts 2 h after administration. Only cells next to lysozyme+ Paneth cells were quantified as CBC stem cells. Crypt cells that were not touching lysozyme+ cells were quantified as transit-amplifying (TA) cells (n = 5 mice per group). Representative image of crypt stained for EdU (cyan), DAPI (nuclei, blue), lysozyme (white) and E-cadherin (red). Scale bar, 20 μm. h, Quantification of Ki67+ cells in human ileal biopsies. Cells at the crypt bottom with elongated nuclei next to postmitotic Paneth cells were counted as CBCs. Cells not at the crypt base were considered transit-amplifying cells. (n = 6 for 20–25-year-old donors, n = 10 for ≥ 75-year-old donors). i, Representative gating of Lgr5hi, Lgr5med, Lgr5lo, Paneth and enteroendocrine cells (in relation to Fig. 1c). Quantification of enteroendocrine cells (n = 30 young, n = 26 old). For FACS gating strategy, see Supplementary Fig. 1. j, Analysis of human ileal biopsy material for lysozyme+ Paneth cells (n values for analysed samples shown). k, Immunostaining and quantification of Olfm4+ stem and progenitor cells (green background, n = 75 crypts from young and old. Five individuals per age group) and lysozyme+ Paneth cells in jejunal crypts (red background, n = 115 crypts from young and n = 117 crypts from old mice, five individuals per age group). Whiskers plotted according to Tukey’s method. Scale bars, 10 μm. l, Ratio of Lgr5hi stem cells and Lgr5med progenitor cells and ratio of Lgr5hi stem cells and Paneth cells analysed by flow cytometry from isolated crypts (n = 30 young, n = 26 old). Whiskers plotted according to Tukey’s method. m, Regenerative growth of young Lgr5hi stem cells co-cultured with young or old Paneth cells. Quantification at day 8–11 (n = 6). Representative images are from day 8. Scale bar, 100 μm. Student’s paired t-test. n, Long-term clonogenicity of young and old Lgr5hi stem cells co-cultured with young and old Paneth cells. Serially passaged organoids were quantified 21 days after initial plating (n = 14 mice per age group). Combinations compared to average of young Lgr5hi cells co-cultured with young and old Paneth cells. o, Fourteen-day co-culture of Paneth cells from tdTomato-expressing mouse (R26-mTmG) with Lgr5hi stem cells from Lgr5-eGFP-IRES-creERT2 mouse show long-term niche interactions in organoid culture. Scale bar, 100 μm. Similar results were seen in three replicate wells from co-cultures of the same mice. Y, mice between 3 and 9 months of age; O, mice over 24 months of age in all experiments. In box plots, unless otherwise indicated, the line represents median, the box shows interquartile range and whiskers represent the range. All other data are mean ± s.d.; two-tailed unpaired Student’s t-test; exact P values shown in corresponding panels.
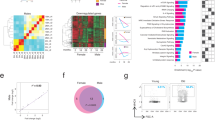
Extended Data Fig. 2 Characterization of gene expression in old Paneth and ISCs.
a, Venn diagram of gene-expression changes in old Paneth cells. (n = 5 mice in old, n = 4 mice in young) b, List of Gene Ontology (GO) terms with the highest enrichment among genes deregulated in old Paneth cells. Fisher’s exact test, no correction for multiple testing. c, Expression of stem-cell-maintaining factors Wnt3 and Egf, and of Notum and Bst1 in old Paneth cells (RNA sequencing (RNA-seq)). Values show fold change in comparison to young Paneth cells. (n = 5 mice in old, n = 4 mice in young). d, Gene editing of Bst1 confirmed by PCR strategy with primers flanking the editing site (191-bp product) and hitting the edited site (89-bp product). Representative agarose gel image is shown. Experiment repeated once to validate the organoid line used in e. e, Regenerative growth of Bst-1 knockout intestinal organoids. Organoids were quantified two days after subculturing (n = 5 repeated experiments with the same organoid line). f, Venn diagram of gene-expression changes in old Lgr5hi stem cells. GSEA preranked analysis of old versus young Lgr5hi stem cells for the gene list ‘KEGG WNT signalling pathway’. Nominal P value is shown (n = 3 mice per age group). g, RNA-scope for NOTUM mRNA (brown) in human jejunal section. Expression seen exclusively in Paneth cells (arrowheads and inset). Experiment repeated twice with similar results in independent samples. h, Expression of human NOTUM and LGR5 from terminal ileal samples of GTEx Consortium (n = 51 sex-matched samples). Expression range is divided into three equal-sized tertiles. In box plots, unless otherwise indicated, the line represents median, the box shows interquartile range and whiskers represent the range. All other data are mean ± s.d.; two-tailed unpaired Student’s t-test; exact P values shown in corresponding panels. For gel source data, see Supplementary Fig. 3.
Extended Data Fig. 3 Wnt ligands increase regenerative capacity of ISCs.
a, Distribution of organoid size on day 5 in ENR + 100 ng ml−1 Wnt3A ± 1μg ml−1 recombinant Notum (n = 50 organoids for Notum-treated (red), n = 38 organoids for untreated (black)). b, Area of colonies from sorted Lgr5hi stem cells from young and old mice (n = 3 mice per age group). Area quantified at day 7. c, Organoid-forming capacity of crypts from young and old mice treated with 100 ng ml−1 Wnt3A. Starting frequency was quantified on day 2 and is represented relative to untreated control (n = 10 mice per age group). d, Primary and secondary regenerative growth of young and old organoids treated with or without 100 ng ml−1 Wnt3A for the first 2 days of culture. Primary organoids were quantified at day 6 and secondary organoids two days after subculturing. Data are represented relative to untreated control (n = 9 mice per age group). e, Organoid-forming capacity of isolated crypts from young mice treated with or without 1 μg ml−1 recombinant Notum (n = 3 mice). f, Primary regenerative growth of organoids from young mice treated with or without 1 μg ml−1 recombinant Notum, quantified on day 6 (n = 3 mice). g, Secondary regenerative growth of organoids from young mice treated with or without 1 μg ml−1 recombinant Notum, quantified on day 2 after subculture (n = 3 mice). h, Organoid-forming capacity of isolated crypts at day 2 from young mice treated with Porcupine inhibitor IWP-237 (n = 3 mice). i, Primary regenerative growth of organoids treated with IWP-2 for the first two days of culture. Organoids were quantified on day 6 (n = 3 mice). j, Flow cytometry analysis of cellular frequencies from Lgr5–eGFP organoids two days after treatment with IWP-2 (n = 4 mice). Y, mice between 3 and 9 months of age; O, mice over 24 months of age in all experiments. In box plots, unless otherwise indicated, the line represents median, the box shows interquartile range and whiskers represent the range. All other data are mean ± s.d.; two-tailed unpaired Student’s t-test; exact P values shown in corresponding panels.
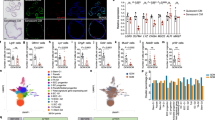
Extended Data Fig. 4 Increased mTORC1 activity in old Paneth cells but not in ISCs.
a, GSEA analysis for gene list ‘Hallmark mTORC1’ (for statistics, see ‘RNA sequencing and data processing’ in Methods). Nominal P value is shown (n = 5 mice in old, n = 4 mice in young). b, Immunohistochemical staining of pS6 (Ser240/244) at mouse jejunal crypt. pS6+ Paneth cells at the crypt bottom are separated by pS6− CBCs. Scale bar, 25 μm. Experiment was repeated for 14 mice with similar results. c, Quantification of pS6+ cells in jejunal crypts (n = 7 mice per age group). d, Isolated Paneth cells from young and old mice, stained with pS6 antibody (red), DAPI (nuclei, blue). Scale bar, 10 μm. Representative image from two independent experiments. e, Left, immunofluorescent image of isolated crypt stained with pS6 antibody (red), Lgr5–eGFP (green) and DAPI (nuclei, blue). Scale bar, 10 μm. Representative of two independent experiments. Right, immunoblots of pS6 and pS6K from isolated crypts of young and old mice, and densitometric quantification (ratio to actin) (n = 14 mice per age group). An outlier (red) deviating >2 s.d. was removed from the analysis. f, Isolated Lgr5hi stem cells (eGFP, green) from young and old mice, stained with pS6 antibody (red), phalloidin (F-actin, white), DAPI (nuclei, blue). Cells were distributed to pS6hi (cells with higher than mean pS6 intensity) or pS6lo (lower than mean pS6 intensity) categories. n = 3 independent experiments. g, Distribution of pS6 intensity in isolated Lgr5hi cells from young and old mice (n = 3 mice per age group, number of cells analysed shown above the corresponding box and whisker plots). h, Mouse weights (n = 25 for young female, n = 26 for old female, n = 20 for young male, n = 19 for old male). Whiskers plotted according to Tukey’s method. Y, mice between 3 and 9 months of age; O, mice over 24 months of age in all experiments. In box plots, unless otherwise indicated, the line represents median, the box shows interquartile range and whiskers represent the range. All other data are mean ± s.d.; two-tailed unpaired Student’s t-test; exact P values shown in corresponding panels. For gel source data, see Supplementary Fig. 3.
Extended Data Fig. 5 Inhibiting mTORC1 activity in old mice restores intestinal regenerative capacity.
a, Organoid-forming capacity and survival of subcultured intestinal organoids treated with rapamycin. Crypts were either treated continuously for four days (2 nM) or with a two-day pulse (2 nM pulse, 10 nM pulse) followed by two days in normal medium before subculturing and quantification (n = 3). b, Regenerative growth of organoids from young and old mice treated with 2 nM rapamycin for 2 days ex vivo. Crypt number was scored six to seven days after treatment from secondary subcultures (two days after passage) (n = 5 mice per age group). Student’s paired t-test. Representative images are from subcultures on day 2. Scale bar, 100 μm. c, Weight of mice receiving daily injections of rapamycin (4 mg kg−1) or vehicle (n = 5 mice per group). Daily data points represent median (circles) and interquartile range (dashed line). d, Immunoblots of pS6 from isolated crypts of vehicle (V)- or rapamycin (R)-treated young and old mice t (n = 4 mice per group). e, Organoid-forming capacity of isolated crypts from old mice treated with vehicle or rapamycin (n = 4 mice per group). f, Primary regenerative growth of organoids from old mice treated with vehicle or rapamycin (n = 4 mice per group). g, Organoid-forming capacity of young Lgr5hi stem cells co-cultured with Paneth cells isolated from young or old mice treated with vehicle or rapamycin (n = 4 mice per group). Combinations compared to average of co-cultures with young vehicle- and old rapamycin-treated Paneth cells. h, Clonogenic growth of Lgr5hi stem cells from young or old mice treated with vehicle or rapamycin (n = 4 mice per group); colonies quantified at day 7. i, RT–qPCR analysis of relative Wnt2b, Wnt5a, Wnt4, Wnt3 and Lgr5 expression from full jejunal samples of old mice treated with rapamycin. Values show fold change (expressed in log2) in comparison to old vehicle treated (n values of mice analysed shown). Data are mean ± s.e.m. j, RT–qPCR analysis of relative Notum and Bst1 expression from crypts of old mice treated with rapamycin. Values show fold change (expressed in log2) in comparison to old vehicle-treated (n = 3 mice per group). Data are mean ± s.e.m. k, Immunoblots of pS6, S6 and H3 from isolated Epcam+ cells of wild type (Tsc1WT) and Tsc1 knockout (Tsc1Δ) epithelium (n = 3 mice per group). l, Quantification of RNA-scope for Notum mRNA in wild type (Tsc1(WT)) and Tsc1 knockout (Tsc1(Δ)) ileal crypts (n = 6 mice for Tsc1(WT) and 5 mice for Tsc1(Δ)). An outlier (red) deviating >2 s.d was removed from the analysis. Representative images of crypts used in quantifications with Notum mRNA (brown) in Paneth cells (inset). m, Organoid-forming capacity of isolated crypts from Tsc1(WT and Tsc1(Δ) epithelium. Quantification was done on day 8. Y, mice between 3 and 9 months of age; O, mice over 24 months of age in all experiments. For gel source data, see Supplementary Fig. 3.
Extended Data Fig. 6 Decreased PPAR activity in aged intestine.
a, GSEA analysis for ‘Biocarta PPARa’ and ‘PPARd’ gene sets (for statistics, see ‘RNA sequencing and data processing’ in Methods). Nominal P value is shown (n = 5 mice in old, n = 4 mice in young). b, Schematic of the putative PPARα-binding site on the mouse and human NOTUM genes found with DECODE. Mouse sequence shown. Bottom, score for the discovered site using JASPAR matrix models for mouse PPAR-response element (PPRE). PPARG; RXRA motif was used. c, FACS analysis of cell populations in primary organoids treated for three days with DMSO, CHIR99021 or GW6471 (n = 6 mice for DMSO, n = 5 mice for CHIR99021 and GW6471). Ratios of Lgr5hi to Paneth cells, and Lgr5hi to Lgr5lo cells from the same analysis. Mean CD24 expression of live Epcam+ cells are also shown. d, Representative images of mouse intestinal organoids treated for 4 days with DMSO, 5 μM GW6471 or 5 μM GW6471 + 100 ng ml−1 Wnt3A. Arrowheads indicate surviving, and red asterisks collapsed, organoids. Scale bar, 100 μm. Experiment was repeated four times with similar results. Unless otherwise indicated, data are mean ± s.d.; two-tailed unpaired Student’s t-test; exact P values shown in corresponding panels.
Extended Data Fig. 7 Notum regulates intestinal stem cell function.
a, Notum gene targeting. Schematic represents sites of genome editing. Gene editing was confirmed by PCR with primers flanking the editing site (174 bp product for Notum KO1 and 294 bp product for Notum KO2) and hitting the edited site (84 bp product for Notum KO1 and 188 bp product for Notum KO2). Representative agarose gel images from two independent experiments with similar results are shown. b, Regenerative growth of Notum knockout organoids. De novo crypt domains were quantified two days after subculture (n = 5 repeated experiments with the same organoid lines). Representative images of organoids two days after subculture are shown. Scale bar, 100 μm. c, Schematic presenting in vivo competition assay of gene-edited organoid growth by orthotopic transplantation to immunodeficient Rag2−/− mice. Representative colonoscopy, necroscopy and histology images used for assay quantification (n = 8 mice transplanted). Scale bars, 1 mm for necroscopy and 200 μm for histology. d, Representative images of CRISPR-targeted young and old organoids two days after subculturing (n = 4 mice per group). Scale bar, 100 μm. e, Relative Notum expression in organoids with synergistic activator mediator complex (SAM) targeted to Notum promoter (dA Notum) grown for two days in ENR medium. Three independent experiments; relative to control (dA Tom). f, Quantification and representative images of day-5 colonies formed by isolated CD24medSSClo cells from Notum activator (dA Notum) and control (dA Tom) organoids. Scale bar, 100 μm. n = 4 repeated experiments with the same organoid line. g, RT–qPCR analysis of relative Axin2 and Lgr5 expression in CD24medSSClo cells sorted from Notum activator (dA Notum) organoids. Values show fold change (expressed in log2) in comparison to control (dATom) (n = 3 replicate wells per organoid line). In box plots, unless otherwise indicated, the line represents median, the box shows interquartile range and whiskers represent the range. All other data are mean ± s.e.m.; two-tailed unpaired Student’s t-test; exact P values shown in corresponding panels. For gel source data see Supplementary Fig. 3.
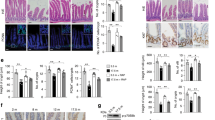
Extended Data Fig. 8 Notum inhibitor ABC99 prevents Wnt inactivation.
a, Flow cytometry analysis of cell populations in primary organoids treated for eight days with 500 nM ABC99 (n = 3 mice) relative to DMSO control. Student’s paired t-test. b, Clonogenic growth of Lgr5hi stem cells on day 5 treated with or without 50 nM ABC99 and/or 500 ng ml−1 recombinant Notum (two independent experiments with similar results, one experiment with three replicate wells shown). c, Relative weight of mice treated with daily injections of ABC99 (10 mg per kg (body weight)) or control (vehicle or ABC101 10 mg per kg (body weight)) (n = 10 mice for young control and young ABC99, n = 8 mice for old control and n = 9 mice for old ABC99). Daily data points represent median (circles) and interquartile range (dashed line). d, Clonogenic growth of young Lgr5hi stem cells co-cultured with young or old Paneth cells from mice treated with ABC99 or control (n values for analysed mice shown). Combinations compared to average of co-cultures with young control (−) and old ABC-treated (+) Paneth cells. Control mice received an equal amount of the inactive analogue ABC101 (yellow circles) or vehicle. e, Representative image of immunofluorescent staining of ileal crypts used for quantification of nuclear β-catenin (white) intensity. Paneth cells (red arrowheads) and CBCs (green arrowheads) were identified by cellular and nuclear (DAPI, blue) morphology. Their nuclear β-catenin levels were compared to transit-amplifying cells (white arrowheads). Scale bar, 20 μm. Experiment was repeated twice with a total of 26 mice all showing strongest nuclear β-catenin at the crypt bottom. f, Immunofluorescent staining of histological sections from old ileum. β-catenin (white), lysozyme (red) and DAPI (nuclei, blue). Scale bar, 10 μm. Quantification of relative nuclear β-catenin intensity of Paneth cells (red arrowhead) (n values for analysed mice shown). For quantification of CBCs (green arrowhead) see Fig. 3d. g, Immunofluorescent staining of histological sections from old ileum. Olfm4, green; EdU, red; DAPI (nuclei), blue. Scale bar, 20 μm. Quantification of EdU+ cellular frequencies within the crypt (n values for analysed mice shown). Y, mice between 3 and 9 months of age; O, mice over 24 months of age in all experiments. In box plots, unless otherwise indicated, the line represents median, the box shows interquartile range and whiskers represent the range. All other data are mean ± s.d.; two-tailed unpaired Student’s t-test; exact P values shown in corresponding panels.
Extended Data Fig. 9 Old intestine recovers poorly from 5-FU-induced damage.
a, Body weights of young and old mice following single injection of 5-FU (100–200 mg kg−1). Two mice per group, body weight relative to day of injection (day 0). b, Relative body weight of young and old mice treated for one week with or without ABC99 followed by single 5-FU (100 mg kg−1) injection (n = 8 mice for young vehicle, old vehicle and young ABC99-treated, n = 10 mice for old ABC99-treated). Daily data points represent median (circles) and interquartile range (dashed line). Daily weight of old ABC99-treated mice were compared to old controls with two-tailed unpaired Student’s t-test; exact P values shown under the corresponding daily weight. Young mice between 3 and 4 months of age; old mice over 20 months of age.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-3 which include the FACS gating strategy (Supplementary Fig. 1), cell purity assessment (Supplementary Fig. 2), uncropped scans of immunoblots and gel electrophoresis (Supplementary Fig. 3) and a full description for Supplementary Table 1.
Supplementary Table 1
Analysis of RNA-sequencing results for young and old Paneth and Lgr5+ ISCs – see Supplementary Information document for full description.
Source data
Rights and permissions
About this article
Cite this article
Pentinmikko, N., Iqbal, S., Mana, M. et al. Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature 571, 398–402 (2019). https://doi.org/10.1038/s41586-019-1383-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1383-0
This article is cited by
-
Intestinal stem cells: guardians of homeostasis in health and aging amid environmental challenges
Experimental & Molecular Medicine (2024)
-
NAD+ dependent UPRmt activation underlies intestinal aging caused by mitochondrial DNA mutations
Nature Communications (2024)
-
The secreted protein Amuc_1409 from Akkermansia muciniphila improves gut health through intestinal stem cell regulation
Nature Communications (2024)
-
A stromal lineage maintains crypt structure and villus homeostasis in the intestinal stem cell niche
BMC Biology (2023)
-
Early weaning causes small intestinal atrophy by inhibiting the activity of intestinal stem cells: involvement of Wnt/β-catenin signaling
Stem Cell Research & Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.