Abstract
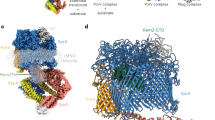
The type 9 secretion system (T9SS) is the protein export pathway of bacteria of the Gram-negative Fibrobacteres–Chlorobi–Bacteroidetes superphylum and is an essential determinant of pathogenicity in severe periodontal disease. The central element of the T9SS is a so-far uncharacterized protein-conducting translocon located in the bacterial outer membrane. Here, using cryo-electron microscopy, we provide structural evidence that the translocon is the T9SS protein SprA. SprA forms an extremely large (36-strand) single polypeptide transmembrane β-barrel. The barrel pore is capped on the extracellular end, but has a lateral opening to the external membrane surface. Structures of SprA bound to different components of the T9SS show that partner proteins control access to the lateral opening and to the periplasmic end of the pore. Our results identify a protein transporter with a distinctive architecture that uses an alternating access mechanism in which the two ends of the protein-conducting channel are open at different times.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM volumes have been deposited in the Electron Microscopy Data Bank under accession codes EMD-0133 and EMD-0134, and the coordinates have been deposited in the Protein Data Bank under accession numbers 6h3i and 6h3j. Source Data for Figs. 1d and 4a, b are available with the online version of the paper.
References
Veith, P. D., Glew, M. D., Gorasia, D. G. & Reynolds, E. C. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol. Microbiol. 106, 35–53 (2017).
Lasica, A. M., Ksiazek, M., Madej, M. & Potempa, J. The type IX secretion system (T9SS): highlights and recent insights into its structure and function. Front. Cell. Infect. Microbiol. 7, 215 (2017).
Li, N. et al. The type IX secretion system is required for virulence of the fish pathogen Flavobacterium columnare. Appl. Environ. Microbiol. 83, e01769-17 (2017).
Pérez-Pascual, D. et al. More than gliding: involvement of GldD and GldG in the virulence of Flavobacterium psychrophilum. Front. Microbiol. 8, 2168 (2017).
Shrivastava, A., Roland, T. & Berg, H. C. The screw-like movement of a gliding bacterium is powered by spiral motion of cell-surface adhesins. Biophys. J. 111, 1008–1013 (2016).
McBride, M. J. & Nakane, D. Flavobacterium gliding motility and the type IX secretion system. Curr. Opin. Microbiol. 28, 72–77 (2015).
Shoji, M. et al. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6, e21372 (2011).
Kulkarni, S. S., Zhu, Y., Brendel, C. J. & McBride, M. J. Diverse C-terminal sequences involved in Flavobacterium johnsoniae protein secretion. J. Bacteriol. 199, e00884-16 (2017).
Kharade, S. S. & McBride, M. J. Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J. Bacteriol. 197, 147–158 (2015).
Heath, J. E. et al. PG1058 is a novel multidomain protein component of the bacterial type IX secretion system. PLoS One 11, e0164313 (2016).
Gorasia, D. G. et al. Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathog. 12, e1005820 (2016).
Leone, P. et al. Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space. Nat. Commun. 9, 429 (2018).
Glew, M. D. et al. PorV is an outer membrane shuttle protein for the type IX secretion system. Sci. Rep. 7, 8790 (2017).
Glew, M. D. et al. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J. Biol. Chem. 287, 24605–24617 (2012).
Gorasia, D. G. et al. Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase-like mechanism. PLoS Pathog. 11, e1005152 (2015).
Nelson, S. S., Glocka, P. P., Agarwal, S., Grimm, D. P. & McBride, M. J. Flavobacterium johnsoniae SprA is a cell surface protein involved in gliding motility. J. Bacteriol. 189, 7145–7150 (2007).
Saiki, K. & Konishi, K. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol. Immunol. 51, 483–491 (2007).
McBride, M. J. & Zhu, Y. Gliding motility and Por secretion system genes are widespread among members of the phylum Bacteroidetes. J. Bacteriol. 195, 270–278 (2013).
Schiffrin, B., Brockwell, D. J. & Radford, S. E. Outer membrane protein folding from an energy landscape perspective. BMC Biol. 15, 123 (2017).
Lasica, A. M. et al. Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci. Rep. 6, 37708 (2016).
Goulas, T. et al. Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Sci. Rep. 5, 11969 (2015).
de Diego, I. et al. Porphyromonas gingivalis virulence factor gingipain RgpB shows a unique zymogenic mechanism for cysteine peptidases. J. Biol. Chem. 288, 14287–14296 (2013).
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C. & Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014).
Dong, H. et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 (2014).
Yan, Z., Yin, M., Xu, D., Zhu, Y. & Li, X. Structural insights into the secretin translocation channel in the type II secretion system. Nat. Struct. Mol. Biol. 24, 177–183 (2017).
Goyal, P. et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516, 250–253 (2014).
Noinaj, N., Gumbart, J. C. & Buchanan, S. K. The β-barrel assembly machinery in motion. Nat. Rev. Microbiol. 15, 197–204 (2017).
Quistgaard, E. M. et al. Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD. BMC Biol. 14, 82 (2016).
van den Berg, B., Black, P. N., Clemons, W. M., Jr & Rapoport, T. A. Crystal structure of the long-chain fatty acid transporter FadL. Science 304, 1506–1509 (2004).
Zgurskaya, H. I., Löpez, C. A. & Gnanakaran, S. Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect. Dis. 1, 512–522 (2015).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
McBride, M. J. & Kempf, M. J. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 178, 583–590 (1996).
Liu, J., McBride, M. J. & Subramaniam, S. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J. Bacteriol. 189, 7503–7506 (2007).
Agarwal, S., Hunnicutt, D. W. & McBride, M. J. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl Acad. Sci. USA 94, 12139–12144 (1997).
Zhu, Y. et al. Genetic analyses unravel the crucial role of a horizontally acquired alginate lyase for brown algal biomass degradation by Zobellia galactanivorans. Environ. Microbiol. 19, 2164–2181 (2017).
Simon, R., Priefer, U. & Puhler, A. A broad host range mobilization system for in vivo genetic engineering—transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1, 784–791 (1983).
Reboul, C. F., Eager, M., Elmlund, D. & Elmlund, H. Single-particle cryo-EM-Improved ab initio 3D reconstruction with SIMPLE/PRIME. Protein Sci. 27, 51–61 (2018).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015).
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protocols 1, 2856–2860 (2006).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Grimm, J. B., Brown, T. A., English, B. P., Lionnet, T. & Lavis, L. D. Synthesis of Janelia Fluor HaloTag and SNAP-Tag ligands and their use in cellular imaging experiments. Methods Mol. Biol. 1663, 179–188 (2017).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Ovesný, M., Křížek, P., Borkovec, J., Svindrych, Z. & Hagen, G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
McBride, M. J. & Braun, T. F. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J. Bacteriol. 186, 2295–2302 (2004).
Acknowledgements
We thank M. McBride and Y. Zhu for providing reagents for the genetic manipulation of F. johnsoniae; A. Shrivastava and H. Berg for advice on measuring gliding motility; L. Lavis for supplying the Janelia Fluor 646 HaloTag ligand; S. Hickman for advice on fluorescence imaging; and O. Meacock and K. Foster for providing additional imaging facilities. We acknowledge the use of Central Oxford Structural Microscopy and Imaging Centre (COSMIC), the Oxford Micron Advanced Imaging Facility, and the Oxford Advanced Proteomics Facility. This work was supported by Wellcome Trust Investigator Awards 107929/Z/15/Z and 100298/Z/12/Z. COSMIC was supported by a Wellcome Trust Collaborative Award 201536/Z/16/Z, the Wolfson Foundation, a Royal Society Wolfson Refurbishment Grant, the John Fell Fund, and the EPA and Cephalosporin Trusts.
Reviewer information
Nature thanks M. McBride and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
F.L. carried out all genetic and biochemical work. J.C.D. collected EM data. J.C.D. and S.M.L. determined the structure. B.C.B. conceived the project. All authors interpreted data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Phenotypic analysis of strains expressing HaloTag and Twin-Strep SprA fusion proteins.
a, b, Immunoblot analysis of HaloTag-SprA (a) and Twin-Strep-SprA (b) expression in whole-cell lysates. Similar data were obtained for three biological repeats. c, Immunoblot detection of levels of the T9SS-dependent chitinase ChiA in culture supernatants. Similar data were obtained for three biological repeats. d, T9SS-dependent spreading (gliding) morphology of colonies on agar. Scale bar, 5 mm. Similar data were obtained for three biological repeats. e, Peptide mass spectrometry of the three highest molecular mass bands in Fig. 1d. Intensity values are normalized to the most abundant SprA peptide detected for each band. Peptide numbering is from the N terminus of the native SprA precursor sequence. For immunoblot source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Experimental quality and resolution estimation of SprA complexes.
a, Representative micrograph of SprA complexes. b, Selected reference-free 2D class averages. c, Gold-standard FSC curves of the final map calculated using a soft-edged mask. d, Local resolution estimates of the final maps. e, Representative density for SprA in the PorV complex (left) and Plug complex (right).
Extended Data Fig. 3 Structural analysis of the SprA complex components.
a, Structural alignment of SprA-bound proteins against the homology models used in initial sequence docking. b, Access routes to the SprA pore viewed from the periplasm (top) or towards the lateral opening (bottom). In the PorV complex two loops involved in coordinating PorV occlude the lateral opening. The step domain is poorly ordered in the Plug complex. For clarity, PorV, Plug, and PPI are not shown. c, The PorV barrel is tilted relative to the SprA barrel. Aromatic residues on the surface of PorV are shown in green spacefill.
Extended Data Fig. 4 Phenotypes of SprA partner deletion strains.
a, Immunoblot analysis of Twin-Strep–SprA levels in whole-cell lysates. Similar data were obtained for three biological repeats. For immunoblot source data, see Supplementary Fig. 1. b, Quantification by liquid chromatography–mass spectrometry of T9SS substrates detected in cell culture supernatants according to CTD type. A detection threshold of more than 1% of the protein abundance in the sprA+ parental strain was applied. c, d, Heat map representations of secreted T9SS substrates from b with type A CTDs (c) or type B CTDs (d). Protein intensities for each protein are normalized to the level detected in the sprA+ parental strain. e, Measurement of vancomycin inhibition zones in a disc diffusion assay. The mean radius of inhibition (red bar) was measured from the disc edge. Error bars represent s.d. (n = 4 for ΔporV and Δplug; n = 5 for other strains) and statistical significance is shown above each measurement set from a one-way ANOVA with post-hoc Dunnett’s test using sprA+ as control group (NS, not significant; ****P < 0.0001). Other comparisons (bracketed) use two-tailed unpaired t-tests; *P = 0.0134, ****P < 0.0001). A control for the DMSO solvent used to dissolve the vancomycin is shown. a–e, All sprA+ strains express the Twin-Strep–SprA fusion protein.
Extended Data Fig. 5 Single-particle cryo-EM image processing workflow for the SprA complexes.
Cryo-EM data sets for SprA complexes in LMNG, in the presence or absence of fluorinated detergents, were combined following 2D classification and subjected to 3D classification against a low-resolution model generated from the fluorinated octyl-maltoside data set. Particle images corresponding to the PorV complex or the Plug complex were then independently refined. A soft mask, generated from these maps, was then used to perform masked refinement against the same particle images, resulting in global map resolutions of 3.5 Å for the PorV complex and 3.7 Å for the Plug complex.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1, which contains the uncropped gel and immunoblot images and Supplementary Table 1, a list of oligonucleotides used in this study
Video 1
Movement of F. johnsoniae cells on glass. F. johnsoniae cells are imaged in real time at 25 °C in tunnel slides containing Motility Medium. The wild-type strain and twinstrep-sprA, halotag-sprA, plug, and ppi mutants display rapid gliding motility. Few sprA or porV cells attach to glass and those that do display limited (sprA) or normal (porV) motility, in agreement with published studies. Scale bar: 5μm. For each strain a representative result is shown. The experiments were each repeated on three independent biological samples with similar results.
Video 2
Live cell fluorescence imaging of HaloTag-SprA. F. johnsoniae cells are imaged in HiLo mode in real time at 25 °C in either tunnel slides containing PY2 medium (for mobile cells) or on agar pads containing PY2 medium (for stationary cells). Scale bar: 5μm Representative results are shown. The experiments were repeated on two independent biological samples with similar results.
Video 3
Architecture of the PorV complex. SprA, blue; PorV, grey; PPI, pink.
Video 4
Architecture of the Plug complex. SprA, blue; Plug, orange; PPI, pink.
Video 5
Changes in SprA conformation between the PorV and Plug complexes. The structures are initially viewed from the periplasmic side of SprA and then from the extracellular side. SprA in chainbows; PorV, Plug, PPI, grey.
Source data
Rights and permissions
About this article
Cite this article
Lauber, F., Deme, J.C., Lea, S.M. et al. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564, 77–82 (2018). https://doi.org/10.1038/s41586-018-0693-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0693-y
Keywords
This article is cited by
-
Structure–function analysis of PorXFj, the PorX homolog from Flavobacterium johnsioniae, suggests a role of the CheY-like domain in type IX secretion motor activity
Scientific Reports (2024)
-
Structural insights into the mechanism of protein transport by the Type 9 Secretion System translocon
Nature Microbiology (2024)
-
Filamentous structures in the cell envelope are associated with bacteroidetes gliding machinery
Communications Biology (2023)
-
Bacterial motility: machinery and mechanisms
Nature Reviews Microbiology (2022)
-
Delivering the pain: an overview of the type III secretion system with special consideration for aquatic pathogens
Veterinary Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.