Abstract
A dominant histopathological feature in neuromuscular diseases, including amyotrophic lateral sclerosis and inclusion body myopathy, is cytoplasmic aggregation of the RNA-binding protein TDP-43. Although rare mutations in TARDBP—the gene that encodes TDP-43—that lead to protein misfolding often cause protein aggregation, most patients do not have any mutations in TARDBP. Therefore, aggregates of wild-type TDP-43 arise in most patients by an unknown mechanism. Here we show that TDP-43 is an essential protein for normal skeletal muscle formation that unexpectedly forms cytoplasmic, amyloid-like oligomeric assemblies, which we call myo-granules, during regeneration of skeletal muscle in mice and humans. Myo-granules bind to mRNAs that encode sarcomeric proteins and are cleared as myofibres mature. Although myo-granules occur during normal skeletal-muscle regeneration, myo-granules can seed TDP-43 amyloid fibrils in vitro and are increased in a mouse model of inclusion body myopathy. Therefore, increased assembly or decreased clearance of functionally normal myo-granules could be the source of cytoplasmic TDP-43 aggregates that commonly occur in neuromuscular disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
eCLIP data are available from the Gene Expression Omnibus (GEO) under accession number GSE104796. Source Data are provided for Figs. 1d, 2c, 3f, h, 4c, 5b, c and Extended Data Figs. 1b, 2c, 3i–k, 4g, 7c, f, h, i, 8c, 9c. All other data supporting the findings of this study are available in the Supplementary Information. Data are available upon request from the corresponding authors.
References
Küsters, B. et al. TDP-43 accumulation is common in myopathies with rimmed vacuoles. Acta Neuropathol. 117, 209–211 (2009).
Weihl, C. C. et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J. Neurol. Neurosurg. Psychiatry 79, 1186–1189 (2008).
Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006).
Renton, A. E., Chiò, A. & Traynor, B. J. State of play in amyotrophic lateral sclerosis genetics. Nat. Neurosci. 17, 17–23 (2014).
Kraemer, B. C. et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 119, 409–419 (2010).
Schmid, B. et al. Loss of ALS-associated TDP-43 in zebrafish causes muscle degeneration, vascular dysfunction, and reduced motor neuron axon outgrowth. Proc. Natl Acad. Sci. USA 110, 4986–4991 (2013).
Diaper, D. C. et al. Drosophila TDP-43 dysfunction in glia and muscle cells cause cytological and behavioural phenotypes that characterize ALS and FTLD. Hum. Mol. Genet. 22, 3883–3893 (2013).
Llamusi, B. et al. Muscleblind, BSF and TBPH are mislocalized in the muscle sarcomere of a Drosophila myotonic dystrophy model. Dis. Model. Mech. 6, 184–196 (2013).
Rodriguez-Ortiz, C. J. et al. Neuronal-specific overexpression of a mutant valosin-containing protein associated with IBMPFD promotes aberrant ubiquitin and TDP-43 accumulation and cognitive dysfunction in transgenic mice. Am. J. Pathol. 183, 504–515 (2013).
Caldwell, C. J., Mattey, D. L. & Weller, R. O. Role of the basement membrane in the regeneration of skeletal muscle. Neuropathol. Appl. Neurobiol. 16, 225–238 (1990).
Hardy, D. et al. Comparative study of injury models for studying muscle regeneration in mice. PLoS ONE 11, e0147198 (2016).
Webster, C., Silberstein, L., Hays, A. P. & Blau, H. M. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell 52, 503–513 (1988).
Johnson, B. S. et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 (2009).
Sangwan, S. et al. Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc. Natl Acad. Sci. USA 114, 8770–8775 (2017).
Kayed, R. et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003).
Van Nostrand, E. L. et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016).
Ayala, Y. M. et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 30, 277–288 (2011).
Polymenidou, M. et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 14, 459–468 (2011).
Tollervey, J. R. et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458 (2011).
Afroz, T. et al. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45 (2017).
Alami, N. H. et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron 81, 536–543 (2014).
Freibaum, B. D., Chitta, R. K., High, A. A. & Taylor, J. P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 (2010).
El Fatimy, R. et al. Tracking the fragile X mental retardation protein in a highly ordered neuronal ribonucleoparticles population: a link between stalled polyribosomes and RNA granules. PLoS Genet. 12, e1006192 (2016).
Taylor, J. P. Multisystem proteinopathy: intersecting genetics in muscle, bone, and brain degeneration. Neurology 85, 658–660 (2015).
Kim, H. J. et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013).
Chiang, P.-M. et al. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc. Natl Acad. Sci. USA 107, 16320–16324 (2010).
Murphy, M. M., Lawson, J. A., Mathew, S. J., Hutcheson, D. A. & Kardon, G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637 (2011).
Salajegheh, M. et al. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve 40, 19–31 (2009).
Custer, S. K., Neumann, M., Lu, H., Wright, A. C. & Taylor, J. P. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum. Mol. Genet. 19, 1741–1755 (2010).
Mompeán, M. et al. Structural evidence of amyloid fibril formation in the putative aggregation domain of TDP-43. J. Phys. Chem. Lett. 6, 2608–2615 (2015).
Chen, A. K.-H. et al. Induction of amyloid fibrils by the C-terminal fragments of TDP-43 in amyotrophic lateral sclerosis. J. Am. Chem. Soc. 132, 1186–1187 (2010).
Igaz, L. M. et al. Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 284, 8516–8524 (2009).
Nishino, I. et al. Distal myopathy with rimmed vacuoles is allelic to hereditary inclusion body myopathy. Neurology 59, 1689–1693 (2002).
Wiesner, D. et al. Reversible induction of TDP-43 granules in cortical neurons after traumatic injury. Exp. Neurol. 299, 15–25 (2018).
Moisse, K. et al. Divergent patterns of cytosolic TDP-43 and neuronal progranulin expression following axotomy: implications for TDP-43 in the physiological response to neuronal injury. Brain Res. 1249, 202–211 (2009).
Wilson, R. S. et al. TDP-43 pathology, cognitive decline, and dementia in old age. JAMA Neurol. 70, 1418–1424 (2013).
Liu, G. et al. Endocytosis regulates TDP-43 toxicity and turnover. Nat. Commun. 8, 2092 (2017).
Laing, N. G. & Nowak, K. J. When contractile proteins go bad: the sarcomere and skeletal muscle disease. BioEssays 27, 809–822 (2005).
Platt, R. J. et al. CRISPR–Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014).
Grimm, J. B. et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015).
Halfmann, R. & Lindquist, S. Screening for amyloid aggregation by semi-denaturing detergent–agarose gel electrophoresis. J. Vis. Exp. 17, 838 (2008).
Fang, Y.-S. et al. Full-length TDP-43 forms toxic amyloid oligomers that are present in frontotemporal lobar dementia-TDP patients. Nat. Commun. 5, 4824 (2014).
Jain, S. et al. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498 (2016).
Winey, M., Meehl, J. B., O’Toole, E. T. & Giddings, T. H. Jr. Conventional transmission electron microscopy. Mol. Biol. Cell 25, 319–323 (2014).
Lovci, M. T. et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 20, 1434–1442 (2013).
Arnauld, S., Bertaux, N., Rigneault, H. & Marguet, D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods 5, 687–694 (2008).
Acknowledgements
We thank J. Dragavon, J. Wei Tay, J. Orth and G. Morgan for help with microscopy; C. Glabe for A11 antibodies, P. Wong for Tardbpflox mice, T. Elston for help with fluorescence-activated cell sorting, T. Lee for help with mass spectrometry; and M. Wicklund, S. Ringel and S. Reed for work with patient samples. The research was supported by NIH-T32GM008497 (J.R.W., E.D.N., T.O.V. and E.L.), NIH-F30NS093682 (J.R.W.), NIH-F30AR068881 (T.O.V.), NIH-GM045443 (R.P.), the Howard Hughes Medical Institute (R.P., D.S.E. and J.P.T.), NIH-R35GM119575 (A.M.J.), Paul O’Hara II Seed Grant from ACS-IRG Grant Program (A.M.J.), University of Colorado Cancer Center Genomics Core (supported by NIH-P30CA46934), NIH-AR049446 and NIH-AR070360 (B.B.O.), Glenn Foundation for Biomedical Research (B.B.O.), Beverly Sears Grant (J.R.W.), NSF MCB 1616265 and NIH NIA AG054022 (D.S.E.) and a Butcher Innovation Award NSF IGERT 1144807 (J.R.W. and T.O.V.).
Reviewer information
Nature thanks A. D. Gitler, E. Olson and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
T.O.V., J.R.W, B.B.O. and R.P. conceived and designed the research, wrote the manuscript and all authors edited drafts. T.O.V., J.R.W., E.L., N.D.B. and O.N.W. performed and analysed mouse regeneration and myotube formation experiments. J.R.W. and E.L. isolated myo-granules. T.E.E., J.R.W. and T.O.V. analysed HaloTag–TDP-43. M.P.H., J.R.W. and T.O.V. performed X-ray diffraction and TEM. E.D.N. and J.R.W. performed eCLIP analysis. T.O.V. and K.A.B. analysed human biopsies. T.O.V. performed VCP experiments. J.R.W. and B.R. performed thioflavin-T assays. E.G., J.S., T.E.L., D.S.E., J.P.T. and A.M.J. provided scientific insights and materials.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
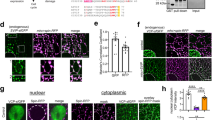
Extended Data Fig. 1 Increased cytosolic TDP-43 during normal skeletal muscle formation.
Related to Fig. 1. a, Nuclear localization of TDP-43 immunofluorescence in C2C12 myoblasts and both nuclear and cytoplasmic localization in C2C12 myotubes differentiated for seven days (n = 3 independent experiment). Myosin heavy chain (MHC) identifies differentiated cells. Scale bars, 25 μm. b, Subcellular fractionation reveals increased cytosolic TDP-43 in differentiating myotubes. Cytosolic (Cyto) myoblasts, 5.0 ± 2.1%; cytosolic myotubes,19.7 ± 3.1%; n = 3 biologically independent experiments that showed similar results, unpaired, two-tailed Student’s t-test, P = 2.0 × 10−3. c, Time course of TDP-43 expression during skeletal-muscle differentiation. n = 3 independent experiments with similar results. Myogenin (MyoG) (magenta) and MHC (green) identify differentiated cells. Nuclei were counterstained with DAPI. Scale bars, 25 μm. d, Top, TDP-43 expression in primary myotubes derived from muscle stem cells that were differentiated in culture for four days. n = 3 independent experiments with similar results. Bottom, images for a secondary-antibody only control. Scale bars, 25 μm. e, Deconvolution microscopy of TDP-43 expression in C2C12 myotubes differentiated for five days. Scale bar, 5 μm. n = 3 independent experiments with similar results. f, CRISPR–Cas9-mediated genomic integration of tetracycline-inducible HaloTag–TDP-43 into the Rosa26 safe-harbour locus in C2C12 myoblasts. A, B and C represent approximate location of primers used in g. g, PCR analyses of gDNA from C2C12 myoblasts for the presence of the HaloTag–TDP-43 construct (top) and integration of the construct into the Rosa26 locus (bottom) using the primers shown in f. n = 3 independent experiments with similar results. Red arrowheads point to the expected PCR product for integration of HaloTag–TDP-43 into Rosa26. Subsequent live-imaging experiments were performed using clones 1 and 4. Non-specific bands are indicated by an asterisk. h, Detection of fluorescently labelled HaloTag–TDP-43 in C2C12 myoblasts following induction resolved on SDS–PAGE. Janelia Fluor 646 (JF646). n = 3 independent experiments with similar results. i, Detection of both HaloTag–TDP-43 and endogenous TDP-43 in selected C2C12 cell clones. n = 3 independent experiments with similar results. j, k, Representative images of individual HaloTag–TDP-43 molecules in a myoblast (j) and a multinucleated myotube (k). Top, start of acquisition (frame 1). Nuclei (Nuc) and cytosolic borders are demarcated by white dotted lines. n = 3 independent experiments with similar results. Bottom, dynamic mapping of single TDP-43 molecule tracks using a multiple target tracing MATLAB script46. Vibrant violet was used to detect myonuclei. Scale bars, 5 μm.
Extended Data Fig. 2 During muscle formation TDP-43 adopts a higher-order state distinct from stress granules.
Related to Fig. 1. a, Secondary antibody-only control for TDP-43 staining of tibialis anterior muscle sections at 5 DPI. Scale bar, 25 μm. n = 5 mice per condition, representative images are shown, all experiments showed similar results. Nuclei were counterstained with DAPI. b, Representative images of TDP-43 and eMHC immunostaining in tibialis anterior muscle sections at 30 DPI; nuclei were counterstained with DAPI. n = 4 mice. Scale bar, 50 μm. c, RIPA–urea assay reveals the presence of an urea-insoluble TDP-43 fraction isolated from C2C12 myotubes that were differentiated for seven days, but not in C2C12 myoblasts. n = 3 independent experiments, each showing similar results, unpaired, two-tailed Student’s t-test, P = 0.0008. GAPDH remains RIPA-soluble in both myoblasts and myotubes. n = 3 independent experiments, each showing similar results, unpaired, two-tailed Student’s t-test, P = 0.7443. d, Higher molecular weight SDS-resistant TDP-43 assemblies were present in differentiating C2C12 myotubes. Protein assemblies resolved by SDD–AGE. n = 3 independent experiments. Pub1Q/N-GFP from yeast forms SDS-resistant assemblies that have a higher molecular weight than TDP-43 assemblies. e, Schematic of the isolation of myo-granules that contain TDP-43 that are formed during skeletal muscle formation. f, Immunoprecipitation (IP) of TDP-43 on Dynabeads (DB) reveals that oligomers isolated from C2C12 myotubes are absent from myoblasts as observed by TEM. n = 3 independent experiments. g, Stress-granule formation in multinucleated myotubes derived from C2C12 cells. Immunofluorescence using antibodies against stress-granule proteins, G3BP1 and PABP1, after NaAsO2 treatment or control conditions for 60 min. n = 3 independent experiments, each showing similar results. Zoom, boxed area shown at higher magnification. Scale bars, 5 μm and 20 μm (insets).
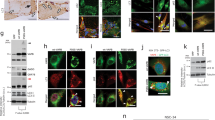
Extended Data Fig. 3 Myo-granules isolated from cells and mice contain TDP-43 and are amyloid-like oligomers.
Related to Fig. 2. a, b, X-ray diffraction of immunoprecipitated myo-granules (right half of a, b) compared to the diffraction of mock IgG immunoprecipitation (left half a) and to the diffraction of super oxide dismutase 1 (SOD1) amyloid oligomers (left half of b). For all diffraction patterns, two rings at approximately 4.8 Å and approximately 10 Å are drawn on the bottom half to highlight the absence of an approximately 4.8 Å reflection in the mock immunoprecipitation and a similar approximately 4.8 Å reflection with the absence of an approximately 10 Å reflection in the SOD1 diffraction. One sample per condition was used. Two diffraction images at different rotations were taken per sample and each image gave similar results. c, d, Complexes that were immunopurified using TDP-43 (c) or A11 (d) were isolated from C2C12 myotubes. Complexes express A11 (c) and TDP-43 (d), whereas immunopurified TDP-43 or A11 myo-granules that were immunostained with secondary antibodies only lack signal. Red, TDP-43 or A11 immunoreactivity. n = 3 independent experiments. Scale bars, 1 μm. e, f, Complexes that were immunopurified using TDP-43 (e) or A11 (f) were isolated from tibialis anterior muscle at 5 DPI. Complexes express A11 (e) and TDP-43 (f), whereas immunopurified TDP-43 or A11 myo-granules immunostained with secondary antibodies only lack signal. Red, TDP-43 or A11 immunoreactivity. n = 3 mice. Scale bars, 0.05 μm. g, TDP-43 immunopurified complexes isolated from an uninjured tibialis anterior muscle (contralateral to the 5 DPI muscle) reveal no complexes with an A11 oligomeric confirmation. n = 3 mice. Scale bars, 0.05 μm. h, A11 immunopurified complexes from an uninjured tibialis anterior muscle (contralateral to the 5 DPI muscle) reveal no complexes containing TDP-43. n = 3 mice. Scale bars, 0.05 μm. i, Dot blot of A11 immunoreactivity in C2C12 cells differentiated into myotubes compared to myoblasts. Quantification reflects fold change in dot blot signal from myoblast to myotube. Data are mean ± s.d., n = 3 independent experiments. j, k, Quantification of the dot blot signal for A11 conformation complexes (j) and TDP-43 conformation complexes (k) during skeletal muscle regeneration at 5 DPI and 10 DPI compared to contralateral uninjured tibialis anterior muscle and normalized to the HRP-only signal. Quantification reflects fold change in dot blot signal. Data are mean ± s.d., n = 3 mice, P values were obtained using unpaired, two-tailed Student’s t-tests.
Extended Data Fig. 4 Myo-granules in skeletal muscle contain TDP-43 and are amyloid-like oligomers.
Related to Fig. 2. a, C2C12 myotubes differentiated for seven days reveal strong A11 immunoreactivity in MHC+ myotubes, but no A11 immunoreactivity in undifferentiated PAX7+ myoblasts. n = 3 independent experiments. Scale bar, 50 μm. b, Muscle stem cells isolated from four-month-old C57/BL6 mice were differentiated in culture for five days and show cytoplasmic and nuclear expression of A11 oligomers. Myotubes express MHC. Scale bar, 50 μm. n = 3 mice. c, Deconvolution microscopy of C2C12 myotubes differentiated for seven days reveal punctate A11 staining in MHC+ myotubes, but no A11 signal was found in undifferentiated myoblasts. Scale bar, 25 μm. n = 3 independent experiments. d, Secondary antibody-only control for A11 staining in tibialis anterior muscle sections at 5 DPI. Nuclei were counterstained with DAPI. Scale bar, 25 μm. n = 4 mice. e, Representative images of A11 and TDP-43 co-localization in tibialis anterior muscle for uninjured muscles, and at 5 DPI and 10 DPI. Scale bars, 25 μm. n = 3 mice. f, Secondary antibody-only control for A11–TDP-43 co-localization in tibialis anterior muscle sections at 5 DPI and 10 DPI shows a lack of signal. Nuclei were counterstained with DAPI. Scale bar, 25 μm. n = 3 mice. g, Quantification of A11 signal intensity in myofibres from e. Unpaired, two-tailed Student’s t-test; comparison between uninjured muscle and 5 DPI, ****P = 4.4 × 10−5; comparison between 5 DPI and 10 DPI, ***P = 4.1 × 10−4; comparison between 10 DPI and uninjured muscle P = 0.024 (P value not shown). n = 3 mice per condition, n = 10 myofibres were averaged per mouse. Data are mean ± s.d. h, Representative deconvolution image of A11 immunoreactivity and eMHC expression in the mouse tibialis anterior myofibres at 5 DPI that were quantified in Fig. 2c. n = 3 mice, each showing similar results. Scale bars, 2 μm and 0.8 μm (inset). i, Proximity ligation assays reveal complexes of TDP-43 and A11 (green) in C2C12 myotubes counterstained with phalloidin (red). A PLA positive control with two antibodies that recognize different epitopes of TDP-43 are positive, whereas complexes are absent if one primary antibody is omitted. n = 3 independent experiments per condition. Ms, mouse; Rb, rabbit.
Extended Data Fig. 5 TDP-43 eCLIP on skeletal muscle myoblasts and myotubes.
Related to Fig. 3. a, RNA immunoprecipitation of C2C12 myotubes, followed by oligo-dT northern blot. Analyses reveal that A11 and TDP-43 associate with poly-A RNA. n = 3 biologically independent samples. b, Schematic of the eCLIP protocol for cultured C2C12 myoblasts and myotubes. c, Immunoprecipitation of TDP-43 complexes used for eCLIP in C2C12 myoblasts. n = 2 biologically independent samples. d, Same as in c, but for C2C12 myotubes. n = 2 biologically independent samples. e, Autoradiogram of 32P-labelled TDP-43–RNA complexes fractionated by PAGE. White boxes indicate the area cut and used for eCLIP library preparation. n = 1 library was prepared per condition. f, Top, scatter plots indicate correlation between significant TDP-43 eCLIP peaks in biological replicates. Scatter plots represent fold enrichment for each region in TDP-43 eCLIP relative to paired size-matched input with significant peaks in red (P ≤ 10−8 over size-matched input). P values for each peak to determine significance were calculated by Yates’ χ2 test (Perl), or Fisher exact test (R computing software) when the expected or observed read number was below five16. For myoblasts, R values were calculated using n = 511,137 non-significant peaks and n = 596 significant peaks. For myotubes, R values were calculated using n = 413,368 non-significant peaks and n = 1,501 significant peaks. Bottom, the UG-rich motif is significantly enriched in clusters from open reading frames and untranslated regions (UTRs). E values were determined using the DREME software tool. g, Irreproducible discovery rate analysis comparing peak fold enrichment across indicated datasets. h, TDP-43 eCLIP reveals that TDP-43 binds to the 3′ UTR of the TDP-43 transcript in myoblasts (top) and myotubes (bottom). n = 3 biologically independent experiments, each showing similar results.
Extended Data Fig. 6 TDP-43 binds to mRNAs that encode sarcomeric proteins during muscle formation.
Related to Fig. 3. a, Myoblast (left), myotube (middle) and shared (right) connectome analysis for all TDP-43 eCLIP peaks (top) and TDP-43 exonic peaks (bottom). b, TDP-43 binds predominantly to exons of protein-coding RNAs in C2C12 myoblasts. c, Peak distribution for significantly enriched TDP-43 peak locations in myoblasts and myotubes across the transcriptome reveal increased exonic and 3′-UTR associations compared to previously identified neuronal TDP-43 peaks18,19. d, Identification of multiple TDP-43-binding sites across and within exons of Ttn. The zoomed region is representative of multiple UG-rich sequences within a single exon. n = 3 biologically independent experiments, each showing similar results.
Extended Data Fig. 7 Myo-granule protein composition.
Related to Fig. 3. a, SDS–PAGE gel stained with SYPRO Ruby reveals enrichment of select proteins during fractionation of total cell lysate (T) from C2C12 myotubes, the enriched fraction (EF) and immunoprecipitation of TDP-43. n = 3 biologically independent experiments, each showing similar results. TDP-43 and IgG control immunoprecipitation experiments are representative of the fractions used for mass spectrometry. b, Venn diagram showing significant overlap between the myo-granule proteome and TDP-43 interactome (previously defined22). The P value was determined using a hypergeometric test. c, Gene Ontology of myo-granules reveals enrichment for processes relating to the localization and translation of RNA. n = 356 proteins, P values were determined using hypergeometric tests with Benjamini–Hochberg false-discovery rate corrections. d, Venn diagram showing significant overlap between myo-granules and neuronal RNA granule proteomes (previously defined23). P value was determined using a hypergeometric test. e, VCP, a top hit in the myo-granule proteome, co-localizes with the cytoplasmic TDP-43 and A11 signals in mouse skeletal muscle at 5 DPI. n = 3 mice. f, The RNA-binding protein HNRNPA2B1 is not associated with the myo-granule proteome and remains localized in myonuclei in injured (5 DPI) and uninjured tibialis anterior muscle. n = 3 mice.
Extended Data Fig. 8 TDP-43 is an essential protein for skeletal muscle formation.
Related to Fig. 3. a, Schematic of the approach used to knockout Tardbp and quantify C2C12 myoblast proliferation. b, Schematic of the isolation and fluorescence-activated cell sorting (FACS) of muscle stem cells from Pax7IREScreTardbpflox/WTRosa26tdTomato and Pax7IREScreTardbpWT/WTRosa26tdTomato mice. More than 125,000 muscle stem cells were collected per mouse from two populations defined in b as TdTom+ and TdTom++. c, Tardbp mRNA expression relative to Gapdh mRNA expression from isolated muscle stem cells from b. n = 4 independent experiments, each a mean of technical triplicates, from n = 2 mice. Unpaired, two-tailed Student’s t-test, *P = 0.0469. Data are mean ± s.d. d, Myofibre feret diameter frequency distribution in uninjured Pax7IREScreTardbpflox/WT mice compared to Pax7IREScreTardbpWT/WT controls. n = 3 mice, 600 myofibres were quantified per condition. e, Quantification of myofibre feret diameter shown in c. In the box plots, the horizontal bars show the mean, boxes show the 25th and 75th percentiles, whiskers show the minimum and maximum, individual myofibres are shown as dots. n = 3 mice, 600 myofibres per condition. Unpaired, two-tailed Student’s t-test, P = 0.5925; ns, not significant. f, Pax7+ muscle stem cell numbers in uninjured Pax7IREScreTardbpflox/WT mice compared to Pax7IREScreTardbpWT/WT controls. n = 3 mice. Unpaired, two-tailed Student’s t-test, P = 0.1963. Data are mean ± s.d. g, Schematic of TDP-43 depletion in Pax7+ muscle stem cells during muscle regeneration in Pax7IREScreTardbpflox/WT and Pax7IREScreTardbpWT/WT mice. Tmx, tamoxifen. h, Quantification of myofibre feret diameters from Fig. 3h at 10 DPI in muscle stem cells from Pax7IREScreTardbpflox/WT mice compared to wild-type controls. In the box plots, the horizontal bars show the mean, boxes show the 25th and 75th percentiles, whiskers show the minimum and maximum, individual myofibres are shown as dots. n = 489 myofibres from n = 3 mice per condition. Unpaired, two-tailed Student’s t-test, ****P = 2.3 × 10−30. i, Similar Pax7+ muscle stem cell numbers at 10 DPI in muscle stem cells from Pax7IREScreTardbpflox/WT haploinsufficient mice compared to wild-type controls. Data are mean ± s.d. from n = 3 mice. Unpaired, two-tailed Student’s t-test, P = 0.89.
Extended Data Fig. 9 Myo-granules that seed amyloid-like fibres are increased in human muscle regeneration and in multisystem proteinopathy.
Related to Figs. 4, 5. a, Representative images of TDP-43 expression (top) and secondary antibody-only control (bottom) in regenerating human skeletal muscle from a patient with necrotizing myopathy. n = 3 independent patient biopsies, each showing similar results. Scale bars, 50 μm. b, Representative tibialis anterior cross-section images of uninjured VCP(A232E) and VCP(WT) mice labelled with EdU after 21 days of EdU treatment in the drinking water to mark division and fusion of muscle stem cells. Laminin identifies myofibres and cells are stained with DAPI to identify nuclei. Arrowheads indicate myofibres with EdU+ centrally located myonuclei. n = 3 mice, each showing similar results. Scale bars, 200 μm and 50 μm (inset). c, Quantification of myofibres with EdU+ centrally located myonuclei in VCP(A232E) and VCP(WT) mice. n = 4 mice, over 1,000 myofibres were quantified per genotype. Data are mean ± s.d. Unpaired, two-tailed Student’s t-test, P = 6.5 × 10−6. d, Representative deconvolution image of A11 and TDP-43 co-localization in a regenerating myofibre from a VCP(A232E) tibialis anterior muscle. n = 3 mice, each showing similar results. Scale bar, 10 μm. e, Secondary antibody-only control of uninjured VCP(A232E) tibialis anterior muscle sections reveals a lack of signal. Nuclei were counterstained with DAPI and myofibres were outlined in white. n = 4 mice, each showing similar results. Scale bar, 25 μm. f, Coomassie-stained recombinant HIS–SUMO–TDP-43 used for thioflavin-T assays resolved by SDS–PAGE. n = 3 biologically independent experiments, each showing similar results. g, Thioflavin-T incorporation reveals thioflavin-T+ amyloid-like fibres for recombinant amyloid-β1–42 and absence of thioflavin-T signal in both the IgG pull-down control and thioflavin-T alone. n = 3 biologically independent experiments, each showing similar results. Scale bars, 10 μm. h, Representative TEM image (zoomed out from Fig. 5e) of thioflavin-T+ (ThT) fibres formed from isolated myo-granules. n = 3 biologically independent experiments. Scale bar, 1 μm.
Extended Data Fig. 10 Myo-granules in normal skeletal muscle regeneration and in disease.
Schematic of TDP-43 oligomerization and aggregation in wild-type, ageing and diseased skeletal muscle myofibres.
Supplementary information
Supplemental Figure 1
Gating strategy for Fluorescence Activated Cell Sorting (FACS) of TdTomato positive muscle stem cells. Muscle stem cells from the hindlimbs of Pax7iresCre;TDP-43flox/wt;ROSA26lox-stop-loxtdTomato and Pax7iresCre;TDP-43wt/wt;ROSA26lox-stop-loxtdTomato mice were isolated by dispersion culture. The cells are genetically labeled for tdTomato (tdTom) and were gated off the known tdTom florescence of mice without tdTom expression in muscle stem cells. Above represents TdTom+ sort and the same strategy was used for the TdTom++ population except TdTom intensity cutoff was 103. Debris was excluded by FSC/SSC, doublets excluded by pulse width vs FSC area and collected cells were DAPI low and tdTom+. DAPI-high positive cells were excluded as they represent dead cells.
Supplementary figures
This figure contains uncropped gel images for Extended Data Fig 1g, h, i; Extended Data Fig 5a, c, d, e; Extended Data Fig 8a.
Supplementary Table 1
Mass spectrometry of myo-granules. Mass spectrometry of isolated TDP-43 containing myo-granules and IgG control IP (n=3 biological independent experiments). TDP-43 interactome defined by Freibaum et al., 2010. RNA-Binding Activity defined by Castello et al., 2010.
Supplementary Table 2
Myo-granule properties identified by mass spectrometry. Protein properties of TDP-43 myo-granules (n=356 proteins) and neuronal RNA granules (n=127 proteins) (El Fatimy et al., 2016). Statistical analysis was performed using hypergeometric distribution to determine chance overlap between two lists. RNA-Binding Activity defined by Castello et al., 2010. Prion-like Domain containing proteins defined using PLAAC (Lancaster et al., 2014) and only proteins with a positive coreSCORE were used in the analysis. Top 400 proteins rich in LARKs (low-complexity, aromatic-rich kinked segments) were searched (Hughes et al., 2018) and used in the analysis.
Supplementary Table 3
Clinical information of patient samples. Clinical features of patients including age, sex, symptoms, symptom duration before biopsy, physical exam features, maximum serum Creatine Kinase level, and clinical diagnoses are indicated. Pathological features including diagnosis, presence of alkaline-phosphatase positive regenerating fibres, myofibre atrophy, myofibre degeneration, and endomysial inflammation are provided.
Supplementary Video 1
HaloTag-TDP-43 in myoblast. Single molecule tracking of HaloTag-TDP-43 in a myoblast.
Supplementary Video 2
HaloTag-TDP-43 in differentiated myotubes. Single molecule tracking of HaloTag-TDP-43 in a differentiated myotube.
Source data
Rights and permissions
About this article
Cite this article
Vogler, T.O., Wheeler, J.R., Nguyen, E.D. et al. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563, 508–513 (2018). https://doi.org/10.1038/s41586-018-0665-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0665-2
Keywords
This article is cited by
-
Nuclear-import receptors as gatekeepers of pathological phase transitions in ALS/FTD
Molecular Neurodegeneration (2024)
-
Neuropathogenesis-on-chips for neurodegenerative diseases
Nature Communications (2024)
-
A guide to membraneless organelles and their various roles in gene regulation
Nature Reviews Molecular Cell Biology (2023)
-
Cryo-EM structure of hnRNPDL-2 fibrils, a functional amyloid associated with limb-girdle muscular dystrophy D3
Nature Communications (2023)
-
Cytotoxic immune cells do not affect TDP-43 and p62 sarcoplasmic aggregation but influence TDP-43 localisation
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.