Abstract
The functions of epithelial tissues are dictated by the types, abundance and distribution of the differentiated cells they contain. Attempts to restore tissue function after damage require knowledge of how physiological tasks are distributed among cell types, and how cell states vary between homeostasis, injury–repair and disease. In the conducting airway, a heterogeneous basal cell population gives rise to specialized luminal cells that perform mucociliary clearance1. Here we perform single-cell profiling of human bronchial epithelial cells and mouse tracheal epithelial cells to obtain a comprehensive census of cell types in the conducting airway and their behaviour in homeostasis and regeneration. Our analysis reveals cell states that represent known and novel cell populations, delineates their heterogeneity and identifies distinct differentiation trajectories during homeostasis and tissue repair. Finally, we identified a novel, rare cell type that we call the ‘pulmonary ionocyte’, which co-expresses FOXI1, multiple subunits of the vacuolar-type H+-ATPase (V-ATPase) and CFTR, the gene that is mutated in cystic fibrosis. Using immunofluorescence, modulation of signalling pathways and electrophysiology, we show that Notch signalling is necessary and FOXI1 expression is sufficient to drive the production of the pulmonary ionocyte, and that the pulmonary ionocyte is a major source of CFTR activity in the conducting airway epithelium.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rock, J. R., Randell, S. H. & Hogan, B. L. M. Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis. Model. Mech. 3, 545–556 (2010).
Hogan, B. L. M. et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–138 (2014).
Hong, K. U., Reynolds, S. D., Watkins, S., Fuchs, E. & Stripp, B. R. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am. J. Physiol. Lung Cell. Mol. Physiol. 286, L643–L649 (2004).
Rock, J. R. et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl Acad. Sci. USA 106, 12771–12775 (2009).
Pardo-Saganta, A. et al. Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell 16, 184–197 (2015).
Van Lommel, A. Pulmonary neuroendocrine cells (PNEC) and neuroepithelial bodies (NEB): chemoreceptors and regulators of lung development. Paediatr. Respir. Rev. 2, 171–176 (2001).
Reid, L. et al. The mysterious pulmonary brush cell: a cell in search of a function. Am. J. Respir. Crit. Care Med. 172, 136–139 (2005).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Gray, T., Guzman, K., Davos, W., Abdullah, L. & Nettesheim, P. Mucocilliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 14, 104–112 (1996).
Tata, P. R. & Rajagopal, J. Plasticity in the lung: making and breaking cell identity. Development 144, 755–766 (2017).
Montoro, D. T. et al. A revised airway hierarchy includes CFTR-expressing ionocytes. Nature https://doi.org/10.1038/s41586-018-0393-7 (2018).
Weinreb, C., Wolock, S. & Klein, A. M. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248 (2017).
Cole, B. B. et al. Tracheal basal cells: a facultative progenitor cell pool. Am. J. Pathol. 177, 362–376 (2010).
Watson, J. K. et al. Clonal dynamics reveal two distinct populations of basal cells in slow-turnover airway epithelium. Cell Reports 12, 90–101 (2015).
Reynolds, S. D., Reynolds, P. R., Pryhuber, G. S., Finder, J. D. & Stripp, B. R. Secretoglobins SCGB3A1 and SCGB3A2 define secretory cell subsets in mouse and human airways. Am. J. Respir. Crit. Care Med. 166, 1498–1509 (2002).
Campell, E. P., Quigley, I. K. & Kintner, C. Foxn4 promotes gene expression required for multiple motile cilia formation. Development 143, 4654–4664 (2016).
Garnett, J. P. et al. Hyperglycaemia and Pseudomonas aeruginosa acidify cystic fibrosis airway surface liquid by elevating epithelial monocarboxylate transporter 2 dependent lactate-H+ secretion. Sci. Rep. 6, 37955 (2016).
Anderson, M. S. & Su, M. A. AIRE expands: new roles in immune tolerance and beyond. Nat. Rev. Immunol. 16, 247–258 (2016).
Galluzzi, L., Kepp, O. & Kroemer, G. Mitochondria: master regulators of danger signalling. Nat. Rev. Mol. Cell Biol. 13, 780–788 (2012).
Vidarsson, H. et al. The forkhead transcription factor Foxi1 is a master regulator of vacuolar H+-ATPase proton pump subunits in the inner ear, kidney and epididymis. PLoS ONE 4, e4471 (2009).
Quigley, I. K., Stubbs, J. L. & Kintner, C. Specification of ion transport cells in the Xenopus larval skin. Development 138, 705–714 (2011).
Spassky, N. & Meunier, A. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 18, 423–436 (2017).
Dubaissi, E. & Papalopulu, N. Embryonic frog epidermis: a model for the study of cell–cell interactions in the development of mucociliary disease. Dis. Model. Mech. 4, 179–192 (2011).
Engelhardt, J. F. et al. Submucosal glands are the predominant site of CFTR expression in the human bronchus. Nat. Genet. 2, 240–248 (1992).
Rock, J. R. et al. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell 8, 639–648 (2011).
Tsao, P. et al. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development 136, 2297–2307 (2009).
Danahay, H. et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Reports 10, 239–252 (2015).
Guseh, J. S. et al. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 136, 1751–1759 (2009).
Kreda, S. M. et al. Characterization of wild-type and ΔF508 cystic fibrosis transmembrane regulator in human respiratory epithelia. Mol. Biol. Cell 16, 2154–2167 (2005).
Li, H., Sheppard, D. N. & Hug, M. J. Transepithelial electrical measurements with the Ussing chamber. J. Cyst. Fibros. 3, 123–126 (2004).
Shah, V. S. et al. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. 351, 503–507 (2016).
Lafkas, D. et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 528, 127–131 (2015).
Li, K. et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J. Biol. Chem. 283, 8046–8054 (2008).
Wu, Y. et al. Therapeutic antibody targeting of individual Notch receptors. Nature 464, 1052–1057 (2010).
Zilionis, R. et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protocols 12, 44–73 (2017).
Zhang, H. M. et al. AnimalTFDB 2.0: A resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 43, D76–D81 (2015).
Weinreb, C., Wolock, S., Tusi, B. K., Socolovsky, M. & Klein, A. M. Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl Acad. Sci. USA 115, E2467–E2476 (2018).
Tusi, B. K. et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60 (2018).
Borthwick, D. W., Shahbazian, M., Krantz, Q. T., Dorin, J. R. & Randell, S. H. Evidence for stem-cell niches in the tracheal epithelium. Am. J. Respir. Cell Mol. Biol. 24, 662–670 (2001).
Acknowledgements
We thank members of the Jaffe and Klein labs, and of the Tissue Repair hub for helpful discussions; C. Weinreb for help with implementing SPRING and data analysis; C. Bauer and K. Feldman for help obtaining human bronchial tissue; P. Capodieci, K. Wetzel and J. Judge for help with tissue processing; K. Mansfield, B. Suchomel and C. Saravanan for help with histology; K. Coote and R. Valdez Misiolek for help with electrophysiology; N. Kirkpatrick, A. Szilvasi and D. Ahern-Ridlon for help with flow cytometry and imaging; J. Reece-Hoyes and S. An for help with lentiviral design; the Single Cell Core Facility at Harvard Medical School for inDrop reagents; the Bauer Core Facility for sequencing; T. Bouwmeester and J. Porter for support and input through the course of this work; R. DeBerardinis for reviewing the manuscript; and B. Ward for editorial assistance. L.W.P. would like to thank R. N. Plasschaert for thoughtful discussion. A.M.K. received support from a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, an Edward Mallinckrodt Jr. Foundation Grant. R.Z. received support from the Lithuanian Education Exchanges Support Foundation. A.M.K. and R.Z. are supported by NCI grant R33CA212697-01.
Reviewer information
Nature thanks I. Amit and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
A.M.K. and A.B.J. conceived the project and performed the initial pilot experiments. L.W.P., R.Z., G.R., A.M.K. and A.B.J. designed experiments and L.W.P. and R.Z. performed most experiments. J.K. and G.R. sequenced cDNA libraries. R.Z, V.S. and A.M.K. performed scRNA-seq analysis. L.W.P. performed histology, lentiviral studies, Notch pathway modulation and electrophysiology. R.C.-W. performed animal work including polidocanol dosing and trachea isolation. L.W.P., R.Z., A.M.K. and A.B.J. wrote the manuscript. All authors read and reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
L.W.P., R.C.-W., J.K., G.R., and A.B.J. are employees of Novartis Institutes for BioMedical Research. V.S. is an employee of Sanofi Aventis. A.M.K. is a founder and SAB member of 1Cell-Bio.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
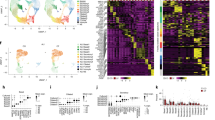
Extended Data Fig. 1 Atlas of transcription factors, surface molecules and kinases enriched in proximal airway lineages of mouse and human.
a, b, Transcription factors, kinases and surface molecules in mouse (a) and human (b) identified among the list of cell-type-specific genes that met the following criteria: significantly enriched in lineage (false discovery rate (FDR) <5%, permutation test), expressed at ≥50 transcripts per million (TPM), expressed in marked lineage at least 1.5× higher than second highest cluster and highest in marked lineage for 4/4 (mouse) or 2/3 (human) biological replicates. c, Pairwise correlation of cell populations identified by single-cell RNA-seq. Genes used for correlation analysis were the 20% most variable genes (identified as described8) of all genes expressed at a level of at least 3 UMI counts in at least 3 cells. Ward’s method was used for hierarchical clustering.
Extended Data Fig. 2 Gene modules identified in mouse tracheal lineages.
Gene modules were identified by selecting variable genes within the given population that were correlated with at least 4 other genes with rank correlation >0.2. a, b, Gene–gene correlation heat map shows 4 gene modules in mouse airway basal cells (a) and 6 gene modules in mouse airway secretory cells (b). SPRING plots show where gene modules are expressed in a given population. Multiple genes are combined in a single signature defined as the mean rank of expression (dense ranking).
Extended Data Fig. 3 Gene modules identified in human bronchial lineages.
a, Two major modules of anti-correlated genes were identified by selecting variable genes within the basal-to-secretory continuum that were correlated with at least 4 other genes with rank correlation >0.12. Genes within each module were then separately considered within basal and secretory cells, keeping genes with a correlation >0.35 with at least 4 other genes. b, c, Gene–gene correlation heat map shows 3 gene modules in human airway basal cells (b) and 4 gene modules in human airway secretory cells (c). SPRING plots show where gene modules are active in a given population. Multiple genes are combined in a single signature defined as the mean rank of expression (dense ranking).
Extended Data Fig. 4 Validation of novel lineages in mouse and human by immunofluorescence.
a, Immunofluorescence in mouse tracheal epithelium for Krt4 (green, arrowheads), Krt5 (basal), Krt8 (luminal), Scgb1a1 (club, secretory) and Foxj1 (ciliated) (n = 3 mice). b, Immunofluorescence in differentiated HBEC cultures for FOXN4 (red, arrows), FOXJ1 (arrowheads mark FOXJ1low cells) and acetylated α-tubulin (cilia) (n = 2 donors). c, Immunofluorescence in HBEC cultures for the ionocyte markers FOXI1, ATP6V1B1 and NGFR (n = 3 donors). Arrowhead shows apical enrichment of ATP6V1B1. Arrows highlight lateral protrusions. Scale bar, 20 μm.
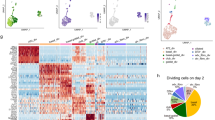
Extended Data Fig. 5 Identification of recovery-specific cell states and population dynamics during regeneration.
a, Cells from uninjured mouse airway do not equally populate all regions of the SPRING plot of all mouse data combined. Each cell from the uninjured condition voted for its 10 nearest neighbours among all mouse cells profiled, and smoothed vote counts were used as a proxy for uninjured cell density on the map (two left-most plots). By visual inspection of the smooth vote distribution a threshold of 25 votes was chosen to binarize regions of the SPRING plot into present versus depleted in uninjured. b, Bar charts representing abundance of rare populations as a fraction of all cells, over time post-injury. Error bars represent the 95% binomial proportion confidence interval (normal approximation). Total number cells = 7,898 from n = 4 mice (uninjured), 898 from n = 1 mouse (1 dpi), 1,964 from n = 1 mouse (2 dpi), 1,082 from n = 1 mouse (3 dpi) and 2,321 from n = 3 mice (7 dpi). c, Bar charts showing the fraction of all cells in each population that express Foxi1 during recovery. Values shown correspond to the fraction of all cells at each time point (cell and mouse numbers as in b above). Error bars defined as in b.
Extended Data Fig. 6 Analysis of basal-to-ciliated differentiation trajectory following injury.
a, Population balance analysis (PBA, see Methods) was used to order 609 cells highlighted in black along the pseudotime of their basal-to-ciliated progression, followed by application of a moving average over a window of 100 cells. The resulting ordering of averaged cells is referred to as the basal-to-ciliated trajectory. PBA requires manually selecting source and sink cells for calculating the pseudotime. b, Heat map of the 1,237 differentially expressed genes along the basal-to-ciliated trajectory (permutation test, FDR <5%, fold-changemax ≥ 2; see Methods). Genes ordered by expression-weighted mean position, defined for an expression time series xt as \(\tau =\frac{\sum _{t}t{x}_{t}}{\sum _{t}{x}_{t}}\). c, Heat map of transcription factors only. Hierarchical clustering revealed six major clusters of correlated genes. Clusters were ordered by mean expression weighted mean position. d, Plots of up to 5 transcription factors sampled from each cluster. The y axis shows the average expression of a gene within the window of 100 cells ± s.e.m. (or +1/[window size] for mean values of zero), normalized to the maximum value. The total trajectory includes 609 cells.
Extended Data Fig. 7 Specification and characterization of FOXI1 lineage in human bronchial epithelium.
a, HBECs were transduced at seeding with GFP or GFP:FOXI1 lentivirus, differentiated and sorted for GFP (shown is representative gating strategy, n = 12). b, Fold change in transduced cells (GFP+) compared to non-transduced cells (GFP−) was determined by RT–qPCR normalized to GAPDH. Pooled data from 2 donors transduced with GFP (n = 7 samples) or GFP:FOXI1 (n = 8 samples) are represented as mean ± s.e.m. P value: FOXI1, 0.001; CFTR, 0.04; ATP6V1B1, 0.006; FOXJ1, 0.01; SCGB1A1 , 0.02; two-tailed t-test. c, Fluorescent in situ hybridization (RNAscope) for FOXI1 and CFTR in HBEC culture transduced with GFP or GFP:FOXI1. Note that while there is an increase in FOXI1/CFTR co-labelled cells, not all FOXI1 cells express CFTR. (arrowheads versus arrows) (n = 2 independent experiments in 2 donors). d, e, Chromogenic in situ hybridization (RNAscope) in primary human bronchial tissue surface epithelium and gland ducts for CFTR and FOXI1 (d) or FOXJ1 (e). Chromogenic signals were split and pseudocoloured to reveal individual channels; inset regions are shown at higher magnification on the right. Note that CFTR is highly enriched in FOXI1+ but not FOXJ1+ cells (n = 1 donor, 5 regions of bronchial tree analysed). Scale bars, 20 μm.
Extended Data Fig. 8 Single-cell RNA-seq analysis of HBECs transduced with GFP and GFP:FOXI1.
a, SPRING plot combining cells transduced with GFP (n = 9,436) or GFP:FOXI1 (n = 10,330), with each of the two conditions highlighted in black (total cells n = 19,766). b, The SLC16A7+ population was identified to be absent in the viral transduction experiment after mapping single-cell transcriptome onto the reference state map. Each cell from the viral transduction experiment voted for its nearest neighbour in the reference experiment. The bar chart on the right shows the average number of votes per cluster. c, Cell states unique to the viral transduction experiment were identified as detailed in Extended Data Fig. 5a. d, Cells representing states also found in the reference experiment (conserved cells) inherited the label of their single nearest neighbour in the reference map. Cells specific to the viral transduction experiment were divided into four clusters by spectral clustering, with their top five enriched genes shown in the top part of the heat map (right). Enrichment of gene g in population i is defined as the fold change in expression of g in i versus the second highest expresser. A pseudo value of 10 TPM was added before calculating the fold change, and only genes expressed at >50 TPM in at least one cluster were considered. The bottom of the heat map shows the top 20 enriched genes identified treating all four transduction-specific states as one population. e, Bar chart showing fold changes in population size following GFP:FOXI1 versus GFP transduction (extension of Fig. 3f). f, Expression of transgene in identified cell populations.
Extended Data Fig. 9 Notch pathway component enrichment in airway lineages.
a, b, SPRING plots show enrichment of Notch pathway components in mouse (a) and human (b) airway lineages. Normalized counts are shown for the Notch ligands JAG1, JAG2 and DLL1, and the Notch receptors NOTCH1, NOTCH2 and NOTCH3. The Notch target gene signature combines HES1, HES5 and NRARP into a single gene signature, defined as the mean expression rank (dense ranking). All gene expression and signature values are smoothed (see Methods for smoothing).
Extended Data Fig. 10 Inhibition of Notch signalling decreases ionocyte markers in HBECs.
a, Expression of Notch target genes and airway lineage markers in cultures treated with 3.3 µM DAPT compared to cultures treated with DMSO. Notch target genes (NRARP P = 0.03, HES5) and secretory cell markers (MUC5B P = 0.001, MUC5AC) are decreased whereas ciliated cell markers (FOXJ1, DNAI2 P = 0.01) and basal cell markers (ITGA6 P = 0.006 and TP63) are increased upon DAPT treatment. Note that ionocyte markers (FOXI1 P = 0.02, CFTR) are also decreased upon DAPT treatment. Two-tailed t-test; n = 8 experiments in 2 donors. b, FOXI1 cell counts in HBEC cultures treated with antibodies that neutralize individual Notch receptors (n = 5 experiments in 2 donors). All data are mean ± s.e.m.
Supplementary information
Supplementary Tables 1-5
This zipped file contains Supplementary Tables 1-5: Supplementary Table 1 Genes enriched in mouse tracheal epithelial cell populations identified by scRNA-seq; Supplementary Table 2 Genes enriched in human bronchial epithelial cell populations identified by scRNA-seq.; Supplementary Table 3 Average gene expression in tracheal epithelial cell populations identified by scRNA-seq in this study; Supplementary Table 4 Differentially expressed genes during basal-to-ciliated differentiation and Supplementary Table 5 Sequences of primers used for inDrops scRNA-seq.
Supplementary Data 1
This file contains a yaml file specifying parameters used with the indrop.py pipeline to process raw mouse data.
Supplementary Data 2
This file contains a yaml file specifying parameters used with the indrop.py pipeline to process raw human data from the reference (untransduced) experiment.
Supplementary Data 3
This file contains a yaml file specifying parameters used with the indrop.py pipeline to process raw human data from the viral transduction experiment.
Source data
Rights and permissions
About this article
Cite this article
Plasschaert, L.W., Žilionis, R., Choo-Wing, R. et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018). https://doi.org/10.1038/s41586-018-0394-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0394-6
This article is cited by
-
Small cells – big issues: biological implications and preclinical advancements in small cell lung cancer
Molecular Cancer (2024)
-
Culture and expansion of murine proximal airway basal stem cells
Stem Cell Research & Therapy (2024)
-
Single-cell division tracing and transcriptomics reveal cell types and differentiation paths in the regenerating lung
Nature Communications (2024)
-
The impacts of active and self-supervised learning on efficient annotation of single-cell expression data
Nature Communications (2024)
-
Dictionary learning for integrative, multimodal and scalable single-cell analysis
Nature Biotechnology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.