Abstract
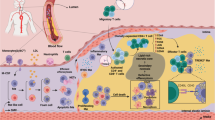
Monocytes and macrophages provide defence against pathogens and danger signals. These cells respond to stimulation in a fast and stimulus-specific manner by utilizing complex cascaded activation by lineage-determining and signal-dependent transcription factors. The complexity of the functional response is determined by interactions between triggered transcription factors and depends on the microenvironment and interdependent signalling cascades. Dysregulation of macrophage phenotypes is a major driver of various diseases such as atherosclerosis, rheumatoid arthritis and type 2 diabetes mellitus. Furthermore, exposure of monocytes, which are macrophage precursor cells, to certain stimuli can lead to a hypo-inflammatory tolerized phenotype or a hyper-inflammatory trained phenotype in a macrophage. In atherosclerosis, macrophages and monocytes are exposed to inflammatory cytokines, oxidized lipids, cholesterol crystals and other factors. All these stimuli induce not only a specific transcriptional response but also interact extensively, leading to transcriptional and epigenetic heterogeneity of macrophages in atherosclerotic plaques. Targeting the epigenetic landscape of plaque macrophages can be a powerful therapeutic tool to modulate pro-atherogenic phenotypes and reduce the rate of plaque formation. In this Review, we discuss the emerging role of transcription factors and epigenetic remodelling in macrophages in the context of atherosclerosis and inflammation, and provide a comprehensive overview of epigenetic enzymes and transcription factors that are involved in macrophage activation.
Key points
Atherosclerotic plaques contain a complex environment with different activation signals that result in intraplaque macrophage heterogeneity.
Plasticity of macrophage phenotype is modulated by an interplay between transcription factors and epigenetic enzymes.
Signalling pathways in inflammation have an extensive molecular crosstalk.
Modulating transcription factor activity is a promising therapeutic target for atherosclerosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Woollard, K. J. & Geissmann, F. Monocytes in atherosclerosis: subsets and functions. Nat. Rev. Cardiol. 7, 77–86 (2010).
Gordon, S. & Plüddemann, A. Tissue macrophages: heterogeneity and functions. BMC Biol. 15, 53 (2017).
Ginhoux, F., Schultze, J. L., Murray, P. J., Ochando, J. & Biswas, S. K. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 17, 34–40 (2016).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 (2014).
Colin, S., Chinetti-Gbaguidi, G. & Staels, B. Macrophage phenotypes in atherosclerosis. Immunol. Rev. 262, 153–166 (2014).
Schultze, J. L., Schmieder, A. & Goerdt, S. Macrophage activation in human diseases. Semin. Immunol. 27, 249–256 (2015).
Moore, K. J., Sheedy, F. J. & Fisher, E. A. Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13, 709–721 (2013).
Cochain, C. et al. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ. Res. 122, 1661–1674 (2018).
Glass, C. K. & Natoli, G. Molecular control of activation and priming in macrophages. Nat. Immunol. 17, 26–33 (2016).
Talbert, P. B. & Henikoff, S. Histone variants — ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275 (2010).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Hoeksema, M. A. & de Winther, M. P. J. Epigenetic regulation of monocyte and macrophage function. Antioxid. Redox Signal. 25, 758–774 (2016).
Zaret, K. S. & Mango, S. E. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr. Opin. Genet. Dev. 37, 76–81 (2016).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Schmidt, S. V. et al. The transcriptional regulator network of human inflammatory macrophages is defined by open chromatin. Cell Res. 26, 151–170 (2016).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Javierre, B. M. et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384.e19 (2016).
Phanstiel, D. H. et al. Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol. Cell 67, 1037–1048.e6 (2017).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Schübeler, D. Function and information content of DNA methylation. Nature 517, 321–326 (2015).
Bock, C. et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol. Cell 47, 633–647 (2012).
Vento-Tormo, R. et al. IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. 17, 4 (2016).
Novakovic, B. et al. β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368.e14 (2016).
Dekkers, K. F., Neele, A. E., Jukema, J. W., Heijmans, B. T. & de Winther, M. P. J. Human monocyte-to-macrophage differentiation involves highly localized gain and loss of DNA methylation at transcription factor binding sites. Epigenetics Chromatin 12, 34 (2019).
Katayama, S. et al. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566 (2005).
Li, H., Jiang, T., Li, M.-Q., Zheng, X.-L. & Zhao, G.-J. Transcriptional regulation of macrophages polarization by microRNAs. Front. Immunol. 9, 3–12 (2018).
Zhang, Z., Salisbury, D. & Sallam, T. Long noncoding RNAs in atherosclerosis: JACC review topic of the week. J. Am. Coll. Cardiol. 72, 2380–2390 (2018).
Biswas, S. K. & Lopez-Collazo, E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30, 475–487 (2009).
Foster, S. L., Hargreaves, D. C. & Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007).
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010).
Arts, R. J. W., Joosten, L. A. B. & Netea, M. G. The potential role of trained immunity in autoimmune and autoinflammatory disorders. Front. Immunol. 9, 298 (2018).
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Cheng, S. C. et al. mTOR- and HIF-1-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Bekkering, S. et al. Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arterioscler. Thromb. Vasc. Biol. 34, 1731–1738 (2014).
Arts, R. J. W. et al. BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe 23, 89–100.e5 (2018).
Ifrim, D. C. et al. Trained immunity or tolerance: opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Clin. Vaccine Immunol. 21, 534–545 (2014).
Yoshida, K. et al. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nat. Immunol. 16, 1034–1043 (2015).
Mitroulis, I. et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 172, 147–155.e12 (2018).
Christ, A. et al. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 172, 162–168.e14 (2018).
Seijkens, T. et al. Hypercholesterolemia-induced priming of hematopoietic stem and progenitor cells aggravates atherosclerosis. FASEB J. 28, 2202–2213 (2014).
Vierbuchen, T. et al. AP-1 transcription factors and the BAF complex mediate signal-dependent enhancer selection. Mol. Cell. 68, 1067–1082.e12 (2017).
Herrington, W., Lacey, B., Sherliker, P., Armitage, J. & Lewington, S. Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ. Res. 118, 535–546 (2016).
GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1151–1210 (2017).
Winkels, H. et al. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ. Res. 122, 1675–1688 (2018).
Stöger, J. L. et al. Distribution of macrophage polarization markers in human atherosclerosis. Atherosclerosis 225, 461–468 (2012).
Martinez, F. O., Sica, A., Mantovani, A. & Locati, M. Macrophage activation and polarization. Front. Biosci. 13, 453–461 (2008).
Benoit, M., Desnues, B. & Mege, J. L. Macrophage polarization in bacterial infections. J. Immunol. 181, 3733–3739 (2008).
Gordon, S. & Martinez, F. O. Alternative activation of macrophages: mechanism and functions. Immunity 32, (593–604 (2010).
Müller, U. et al. IL-13 induces disease-promoting type 2 cytokines, alternatively activated macrophages and allergic inflammation during pulmonary infection of mice with Cryptococcus neoformans. J. Immunol. 179, 5367–5377 (2007).
Spiller, K. L. et al. Differential gene expression in human, murine, and cell line-derived macrophages upon polarization. Exp. Cell Res. 347, 1–13 (2016).
Tugal, D., Liao, X. & Jain, M. K. Transcriptional control of macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 33, 1135–1144 (2013).
Oh, K.-S. et al. Dual roles for ikaros in regulation of macrophage chromatin state and inflammatory gene expression. J. Immunol. 201, 757–771 (2018).
Liu, T., Zhang, L., Joo, D. & Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target Ther. 2, 17023 (2017).
Dorrington, M. G. & Fraser, I. D. C. NF-κB signaling in macrophages: dynamics, crosstalk, and signal integration. Front. Immunol. 10, 705 (2019).
Ghisletti, S. et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 (2010).
Günthner, R. & Anders, H.-J. Interferon-regulatory factors determine macrophage phenotype polarization. Mediators Inflamm. 2013, 731023 (2013).
Mancino, A. et al. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 29, 394–408 (2015).
Saliba, D. G. et al. IRF5:RelA interaction targets inflammatory genes in macrophages. Cell Rep. 8, 1308–1317 (2014).
Szanto, A. et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity 33, 699–712 (2010).
Czimmerer, Z. et al. The transcription factor stat6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity 48, 75–76 (2018).
Chistiakov, D. A., Melnichenko, A. A., Myasoedova, V. A., Grechko, A. V. & Orekhov, A. N. Mechanisms of foam cell formation in atherosclerosis. J. Mol. Med. 95, 1153–1165 (2017).
Tangirala, R. K. et al. Identification of macrophage liver X receptors as inhibitors of atherosclerosis. Proc. Natl Acad. Sci. USA 99, 11896–11901 (2002).
Kim, K. et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ. Res. 123, 1127–1142 (2018).
Wolfs, I. M., Donners, M. M. & de Winther, M. P. Differentiation factors and cytokines in the atherosclerotic plaque micro-environment as a trigger for macrophage polarisation. Thromb. Haemost. 106, 763–771 (2011).
Tall, A. R. & Yvan-Charvet, L. Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 15, 104–116 (2015).
Ogawa, S. et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 122, 707–721 (2005).
Ghisletti, S. et al. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol. Cell 25, 57–70 (2007).
Ghisletti, S. et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 23, 681–693 (2009).
Daniel, B. et al. The IL-4/STAT6/PPARγ signaling axis is driving the expansion of the RXR heterodimer cistrome, providing complex ligand responsiveness in macrophages. Nucleic Acids Res. 46, 4425–4439 (2018).
Daniel, B. et al. The nuclear receptor PPARγ; controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity 49, 615–616 (2018).
Gold, E. S. et al. ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. J. Exp. Med. 209, 807–817 (2012).
De Nardo, D. et al. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat. Immunol. 15, 152–160 (2014).
Li, Z. et al. Krüppel-like factor 4 regulation of cholesterol-25-hydroxylase and liver x receptor mitigates atherosclerosis susceptibility. Circulation 136, 1315–1330 (2017).
Hamada, M. et al. MafB promotes atherosclerosis by inhibiting foam-cell apoptosis. nature communications. Nat. Commun. 5, 3147 (2014).
Hasegawa, H. et al. The role of macrophage transcription factor MafB in atherosclerotic plaque stability. Atherosclerosis 250, 133–143 (2016).
Dubland, J. A. & Francis, G. A. So much cholesterol: the unrecognized importance of smooth muscle cells in atherosclerotic foam cell formation. Curr. Opin. Lipidol. 27, 155–161 (2016).
Holycross, B. J., Blank, R. S., Thompson, M. M., Peach, M. J. & Owens, G. K. Platelet-derived growth factor-BB-induced suppression of smooth muscle cell differentiation. Circ. Res. 71, 1525–1532 (1992).
Barrett, T. B. & Benditt, E. P. Platelet-derived growth factor gene expression in human atherosclerotic plaques and normal artery wall. Proc. Natl Acad. Sci. USA 85, 2810–2814 (1988).
Owens, G. K., Kumar, M. S. & Wamhoff, B. R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84, 767–801 (2004).
Williams, K. J. & Tabas, I. The response-to-retention hypothesis of early atherogenesis. Arterioscler. Thromb. Vasc. Biol. 15, 551–561 (1995).
Ma, S., Yang, D., Li, D., Tang, B. & Yang, Y. Oleic acid induces smooth muscle foam cell formation and enhances atherosclerotic lesion development via CD36. Lipids Health Dis. 10, 53 (2011).
Costales, P. et al. K domain CR9 of low density lipoprotein (LDL) receptor-related protein 1 (LRP1) is critical for aggregated LDL-induced foam cell formation from human vascular smooth muscle cells. J. Biol. Chem. 290, 14852–14865 (2015).
Rivera, J. et al. Accumulation of serum lipids by vascular smooth muscle cells involves a macropinocytosis-like uptake pathway and is associated with the downregulation of the ATP-binding cassette transporter A1. Naunyn Schmiedebergs Arch. Pharmacol. 386, 1081–1093 (2013).
Rong, J. X., Shapiro, M., Trogan, E. & Fisher, E. A. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc. Natl Acad. Sci. USA 100, 13531–13536 (2003).
Allahverdian, S., Chehroudi, A. C., McManus, B. M., Abraham, T. & Francis, G. A. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation 129, 1551–1559 (2014).
Shankman, L. S. et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 21, 628–637 (2015).
Deaton, R. A., Gan, Q. & Owens, G. K. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am. J. Physiol. Heart Circ. Physiol. 296, H1027–H1037 (2009).
Kapoor, N. et al. Transcription factors STAT6 and KLF4 implement macrophage polarization via the dual catalytic powers of MCPIP. J. Immunol. 194, 6011–6023 (2015).
Vengrenyuk, Y. et al. Cholesterol loading reprograms the microRNA-143/145-myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arterioscler. Thromb. Vasc. Biol. 35, 535–546 (2015).
Feil, S. et al. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ. Res. 115, 662–667 (2014).
Huff, M. W. & Pickering, J. G. Can a vascular smooth muscle-derived foam-cell really change its spots? Arterioscler. Thromb. Vasc. Biol. 35, 492–495 (2015).
Li, B. et al. Kallistatin inhibits atherosclerotic inflammation by regulating macrophage polarization. Hum. Gene Ther. 30, 339–351 (2019).
Chen, W. et al. Tanshinone IIA harmonizes the crosstalk of autophagy and polarization in macrophages via miR-375/KLF4 pathway to attenuate atherosclerosis. Int. Immunopharmacol. 70, 486–497 (2019).
Tang, R.-Z. et al. DNA methyltransferase 1 and Krüppel-like factor 4 axis regulates macrophage inflammation and atherosclerosis. J. Mol. Cell Cardiol. 128, 11–24 (2019).
Ivashkiv, L. B. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 34, 216–223 (2013).
Mullican, S. E. et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 25, 2480–2488 (2011).
Hoeksema, M. A. et al. Targeting macrophage histone deacetylase 3 stabilizes atherosclerotic lesions. EMBO Mol. Med. 6, 1124–1132 (2014).
You, S.-H. et al. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol. 20, 182–187 (2013).
Li, P. et al. NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell 155, 200–214 (2013).
Cao, Q. et al. Histone deacetylase 9 represses cholesterol efflux and alternatively activated macrophages in atherosclerosis development. Arterioscler. Thromb. Vasc. Biol. 34, 1871–1879 (2014).
Satoh, T. et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 (2010).
Neele, A. E. et al. Macrophage Kdm6b controls the pro-fibrotic transcriptome signature of foam cells. Epigenomics 9, 383–391 (2017).
Neele, A. E. et al. Myeloid Kdm6b deficiency results in advanced atherosclerosis. Atherosclerosis 275, 156–165 (2018).
Hsu, A. T. et al. Epigenetic and transcriptional regulation of IL4-induced CCL17 production in human monocytes and murine macrophages. J. Biol. Chem. 293, 11415–11423 (2018).
Achuthan, A. et al. Granulocyte macrophage colony-stimulating factor induces CCL17 production via IRF4 to mediate inflammation. J. Clin. Invest. 126, 3453–3466 (2016).
Lehtonen, A., Matikainen, S., Miettinen, M. & Julkunen, I. Granulocyte-macrophage colony-stimulating factor (GM-CSF)-induced STAT5 activation and target-gene expression during human monocyte/macrophage differentiation. J. Leukoc. Biol. 71, 511–519 (2002).
Krausgruber, T. et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 12, 231–238 (2011).
Zhang, X. et al. Macrophage/microglial Ezh2 facilitates autoimmune inflammation through inhibition of Socs3. J. Exp. Med. 215, 1365–1382 (2018).
Jaiswal, S. et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017).
Fuster, J. J. et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017).
Sun, J. et al. SIRT1 activation disrupts maintenance of myelodysplastic syndrome stem and progenitor cells by restoring TET2 function. Cell Stem Cell 23, 355–359 (2018).
Stein, S. & Matter, C. M. Protective roles of SIRT1 in atherosclerosis. Cell Cycle 10, 640–647 (2014).
Sano., S. et al. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ. Res. 123, 335–341 (2018).
Li, X. et al. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 17, 806–815 (2016).
Lin, J.-D. et al. Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight 4, e124574 (2019).
Piccolo, V. et al. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol. 18, 530–540 (2017).
Qiao, Y., Kang, K., Giannopoulou, E., Fang, C. & Ivashkiv, L. B. IFN-γ induces histone 3 lysine 27 trimethylation in a small subset of promoters to stably silence gene expression in human macrophages. Cell Rep. 16, 3121–3129 (2016).
Kang, K. et al. Interferon-γ represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity 47, 235–250.e4 (2017).
Eichenfield, D. Z. et al. Tissue damage drives co-localization of NF-κB, Smad3, and Nrf2 to direct Rev-erb sensitive wound repair in mouse macrophages. eLife 5, e13024 (2016).
Park, S. H. et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol. 18, 1104–1116 (2017).
Neele, A. E., Van den Bossche, J., Hoeksema, M. A. & de Winther, M. P. J. Epigenetic pathways in macrophages emerge as novel targets in atherosclerosis. Eur. J. Pharmacol. 763, 79–89 (2015).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Parnas, O. et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell 162, 675–686 (2015).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1857.e17 (2016).
Acknowledgements
The work of M.P.J.de W. and C.K.G. is supported by The Netherlands Heart Foundation (GENIUS (CVON 2011/B019) and GENIUS2 (CVON 2017–20) to M.P.J.de W.); The Netherlands Heart Foundation and Spark-Holding BV (2015B002 to M.P.J.de W.); the European Union (ITN grant EPIMAC to M.P.J.de W.); NIH grants (DK063491, DK091183 and HL088093 to C.K.G.); and Foundation Leducq (LEAN Transatlantic Network Grant to M.P.J.de W. and C.K.G.).
Author information
Authors and Affiliations
Contributions
All the authors researched data for the article, discussed its content, wrote the manuscript and reviewed and edited it before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Transcription factors
-
Proteins with a DNA-binding domain that guide the transcriptional machinery to or block it from regulatory elements such as promoters and enhancers, thereby facilitating the execution of tailored transcriptional programmes.
- Long-range interactions
-
Interactions between regulatory DNA regions that occur over large linear distances, predominantly between enhancers and between enhancers and promoters. These interactions can have a regulatory role.
- Promoters
-
Regulatory DNA regions proximal to a transcription start site. Contain consensus motifs for transcription factors and the transcriptional machinery.
- Enhancers
-
Regulatory DNA regions distal to a transcription start site. Contain transcription factor binding sites that can regulate the transcription of target genes independently of linear distance.
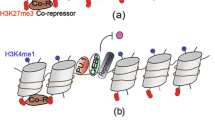
- Histone-modifying enzymes
-
(HMEs). Readers, writers or erasers of chemical histone (tail) modifications.
- Lineage-determining transcription factors
-
(LDTFs). Transcription factors with pioneering (that is, chromatin conformation-changing) capabilities that set up the chromatin for execution of cell type-specific gene programmes.
- Signal-dependent transcription factors
-
(SDTFs). Transcription factors that bind DNA and activate its gene programme in response to an external stimulus.
- CpG islands
-
Genomic regions with a higher-than-average frequency of CpG sites, typically defined as a region of 500–1,500 bp in length, with CpG content >50% and an observed/expected CpG ratio of >0.6; CpG islands often occur in gene regulatory regions, and their DNA methylation status regulates genomic structures and gene transcription.
- Histone marks
-
Chemical groups covalently bound to a specific amino acid residue on (the tail of) a histone protein. Histone marks can change the local chromatin accessibility through electrostatic effects and/or guide the transcription machinery to specific loci in the genome (Box 1).
- Euchromatin
-
A permissive chromatin state in which the chromatin is in an open conformation accessible to the transcription machinery.
- Heterochromatin
-
A repressive chromatin state in which the chromatin is in a closed conformation inaccessible to transcription factors.
- Assay for transposase-accessible chromatin using sequencing
-
(ATAC-seq). Method for genome-wide identification of open chromatin regions with potential regulatory function.
Rights and permissions
About this article
Cite this article
Kuznetsova, T., Prange, K.H.M., Glass, C.K. et al. Transcriptional and epigenetic regulation of macrophages in atherosclerosis. Nat Rev Cardiol 17, 216–228 (2020). https://doi.org/10.1038/s41569-019-0265-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41569-019-0265-3
This article is cited by
-
Advances in secondary prevention mechanisms of macrovascular complications in type 2 diabetes mellitus patients: a comprehensive review
European Journal of Medical Research (2024)
-
YAP represses intestinal inflammation through epigenetic silencing of JMJD3
Clinical Epigenetics (2024)
-
NOS-like activity of CeO2 nanozymes contributes to diminishing the vascular plaques
Journal of Nanobiotechnology (2024)
-
Nobiletin alleviates atherosclerosis by inhibiting lipid uptake via the PPARG/CD36 pathway
Lipids in Health and Disease (2024)
-
Neutrophil extracellular traps: a catalyst for atherosclerosis
Molecular and Cellular Biochemistry (2024)