Abstract
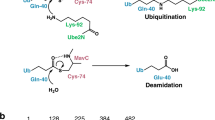
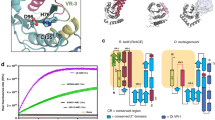
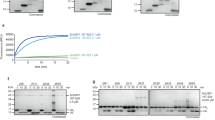
Linear ubiquitin (Ub) chains regulate many cellular processes, including NF-κB immune signalling. Pathogenic bacteria have evolved to secrete effector proteins that harbour deubiquitinase activity into host cells to disrupt host ubiquitination signalling. All previously identified effector deubiquitinases hydrolyse isopeptide-linked polyubiquitin (polyUb). It has been a long-standing question whether bacterial pathogens have evolved an effector deubiquitinase to directly cleave linear Ub chains. In this study, we performed extensive screening of bacterial pathogens and found that Legionella pneumophila—the causative agent of human Legionnaire’s disease—encodes an effector protein, RavD, which harbours deubiquitinase activity exquisitely specific for linear Ub chains. RavD hydrolyses linear Ub chains but not any type of isopeptide-linked polyUb. The crystal structure of RavD with linear diubiquitin reveals that RavD adopts a papain-like fold with a Cys–His–Ser catalytic triad. The Ub-binding surface and specific interacting residues in RavD determine its specificity for Met1 linkages. RavD prevents the accumulation of linear Ub chains on Legionella-containing vacuoles established by the pathogen in host cells to inhibit the NF-κB pathway during infection. This study identified a unique linear Ub chain-specific effector deubiquitinase and indicates its potential application as a tool to dissect linear polyUb-mediated signalling in mammalian cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Coordinates and structure factors for RavD and the RavD-linear diUb complex have been deposited in the Protein Data Bank under accession codes 6NII and 6NJD, respectively. All data generated during this study are included in this published article and its Supplementary Information.
References
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev. Cell Dev. Biol. 22, 159–180 (2006).
Zhou, Y. & Zhu, Y. Diversity of bacterial manipulation of the host ubiquitin pathways. Cell Microbiol. 17, 26–34 (2015).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 (2011).
Tokunaga, F. et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006).
Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011).
Hrdinka, M. & Gyrd-Hansen, M. The Met1-linked ubiquitin machinery: emerging themes of (de)regulation. Mol. Cell 68, 265–280 (2017).
Komander, D., Clague, M. J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
Mevissen, T. E. T. & Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 (2017).
Keusekotten, K. et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326 (2013).
Rivkin, E. et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324 (2013).
Damgaard, R. B. et al. The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell 166, 1215–1230 (2016).
Zhou, Q. et al. Biallelic hypomorphic mutations in a linear deubiquitinase define otulipenia, an early-onset autoinflammatory disease. Proc. Natl Acad. Sci. USA 113, 10127–10132 (2016).
Hubber, A. & Roy, C. R. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283 (2010).
Wan, M., Zhou, Y. & Zhu, Y. Subversion of macrophage functions by bacterial protein toxins and effectors. Curr. Issues Mol. Biol. 25, 61–80 (2018).
Zhou, Y. et al. Nε-fatty acylation of Rho GTPases by a MARTX toxin effector. Science 358, 528–531 (2017).
Cui, J. & Shao, F. Biochemistry and cell signaling taught by bacterial effectors. Trends Biochem. Sci. 36, 532–540 (2011).
Pruneda, J. N. et al. The molecular basis for ubiquitin and ubiquitin-like specificities in bacterial effector proteases. Mol. Cell 63, 261–276 (2016).
Noad, J. et al. LUBAC-synthesized linear ubiquitin chains restrict cytosol-invading bacteria by activating autophagy and NF-κB. Nat. Microbiol. 2, 17063 (2017).
Van Wijk, S. J. L. et al. Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF-κB and restricts bacterial proliferation. Nat. Microbiol. 2, 17066 (2017).
Rytkonen, A. et al. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl Acad. Sci. USA 104, 3502–3507 (2007).
Swanson, M. S. & Hammer, B. K. Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54, 567–613 (2000).
Isberg, R. R., O’Connor, T. J. & Heidtman, M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 (2009).
Zhu, W. et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE 6, e17638 (2011).
Sheedlo, M. J. et al. Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination. Proc. Natl Acad. Sci. USA 112, 15090–15095 (2015).
Hilbi, H., Weber, S. & Finsel, I. Anchors for effectors: subversion of phosphoinositide lipids by Legionella. Front Microbiol 2, 91 (2011).
Haneburger, I. & Hilbi, H. Phosphoinositide lipids and the Legionella pathogen vacuole. Curr. Top. Microbiol. Immunol. 376, 155–173 (2013).
Zhu, M., Shao, F., Innes, R. W., Dixon, J. E. & Xu, Z. The crystal structure of Pseudomonas avirulence protein AvrPphB: a papain-like fold with a distinct substrate-binding site. Proc. Natl Acad. Sci. USA 101, 302–307 (2004).
Shao, F. et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301, 1230–1233 (2003).
Sanada, T. et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature 483, 623–626 (2012).
Fu, P. et al. Complex structure of OspI and Ubc13: the molecular basis of Ubc13 deamidation and convergence of bacterial and host E2 recognition. PLoS Pathog. 9, e1003322 (2013).
Schlieker, C. et al. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25, 677–687 (2007).
Rahighi, S. et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136, 1098–1109 (2009).
Price, C. T., Al-Quadan, T., Santic, M., Rosenshine, I. & Abu Kwaik, Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334, 1553–1557 (2011).
Hsu, F. et al. The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling. Proc. Natl Acad. Sci. USA 111, 10538–10543 (2014).
Pike, C. M., Boyer-Andersen, R., Kinch, L. N., Caplan, J. L. & Neunuebel, M. R. Legionella effector RavD binds phosphatidylinositol-3-phosphate and helps suppress endolysosomal maturation of the Legionella-containing vacuole.J. Biol. Chem. 294, 6405–6415 (2019).
Peltzer, N. et al. LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature 557, 112–117 (2018).
Heger, K. et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature 559, 120–124 (2018).
Takiuchi, T. et al. Suppression of LUBAC-mediated linear ubiquitination by a specific interaction between LUBAC and the deubiquitinases CYLD and OTULIN. Genes Cells 19, 254–272 (2014).
Zinngrebe, J. et al. LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J. Exp. Med. 213, 2671–2689 (2016).
Yin, Q. et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 16, 658–666 (2009).
Stieglitz, B. et al. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature 503, 422–426 (2013).
Doublie, S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530 (1997).
Wang, Z. et al. Automatic crystal centring procedure at the SSRF macromolecular crystallography beamline. J. Synchrotron Radiat. 23, 1323–1332 (2016).
Kabsch, W. XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 (2010).
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D. Biol. Crystallogr. 58, 1772–1779 (2002).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 (1997).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 62, 859–866 (2006).
Acknowledgements
We thank the staff at the beamline BL18U1 of SSRF and the National Center for Protein Science Shanghai for assistance with diffraction data collection, F. Shao, L. Wang, V. Dixit and Z.-Q. Luo for providing bacterial strains and antibodies, and the staff at the core facilities of the Life Sciences Institute Zhejiang University for assistance with SIM. This work was supported by grants from MOST (2017YFA0503900 to Y.Zhu), NSFC (81530068 to Y.Zhu and 81501717 to Y.Zhou), the National High-level Talents Special Support Plan, the leading scientist program of Zhejiang High-level Talents Special Support Plan and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Contributions
Y.Zhu designed the study. M.W. performed the biochemical, cell and infection assays with assistance from Y.Zhou, X.W., C.H., D.X. and Z.W. X.W. determined the structures and generated the mutant strains. M.W., X.W., Y.Zhou and Y.Zhu analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–19, Supplementary Table 1 and uncropped blots
Rights and permissions
About this article
Cite this article
Wan, M., Wang, X., Huang, C. et al. A bacterial effector deubiquitinase specifically hydrolyses linear ubiquitin chains to inhibit host inflammatory signalling. Nat Microbiol 4, 1282–1293 (2019). https://doi.org/10.1038/s41564-019-0454-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0454-1
This article is cited by
-
Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs
Nature Chemical Biology (2024)
-
Functional and structural diversity in deubiquitinases of the Chlamydia-like bacterium Simkania negevensis
Nature Communications (2023)
-
Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nature Communications (2022)
-
The Met1-linked ubiquitin machinery in inflammation and infection
Cell Death & Differentiation (2021)
-
Met1-linked ubiquitin signalling in health and disease: inflammation, immunity, cancer, and beyond
Cell Death & Differentiation (2021)