Abstract
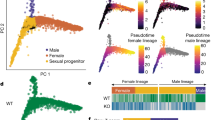
During malaria infection, Plasmodium spp. parasites cyclically invade red blood cells and can follow two different developmental pathways. They can either replicate asexually to sustain the infection, or differentiate into gametocytes, the sexual stage that can be taken up by mosquitoes, ultimately leading to disease transmission. Despite its importance for malaria control, the process of gametocytogenesis remains poorly understood, partially due to the difficulty of generating high numbers of sexually committed parasites in laboratory conditions1. Recently, an apicomplexa-specific transcription factor (AP2-G) was identified as necessary for gametocyte production in multiple Plasmodium species2,3, and suggested to be an epigenetically regulated master switch that initiates gametocytogenesis4,5. Here we show that in a rodent malaria parasite, Plasmodium berghei, conditional overexpression of AP2-G can be used to synchronously convert the great majority of the population into fertile gametocytes. This discovery allowed us to redefine the time frame of sexual commitment, identify a number of putative AP2-G targets and chart the sequence of transcriptional changes through gametocyte development, including the observation that gender-specific transcription occurred within 6 h of induction. These data provide entry points for further detailed characterization of the key process required for malaria transmission.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Meibalan, E. & Marti, M. Biology of malaria transmission. Cold Spring Harb. Perspect. Med. 7, a025452 (2017).
Sinha, A. et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature 507, 253–257 (2014).
Kafsack, B. F. C. et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252 (2014).
Brancucci, N. M. B. et al. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe 16, 165–176 (2014).
Filarsky, M. et al. GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science 359, 1259–1263 (2018).
Brancucci, N. M. B. et al. Lysophosphatidylcholine regulates sexual stage differentiation in the human malaria parasite Plasmodium falciparum. Cell 171, 1532–1544 (2017).
Jullien, N., Sampieri, F., Enjalbert, A. & Herman, J.-P. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 31, e131 (2003).
Albert, H., Dale, E. C., Lee, E. & Ow, D. W. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7, 649–659 (1995).
Mair, G. R. et al. Universal features of post-transcriptional gene regulation are critical for Plasmodium zygote development. PLoS Pathog. 6, e1000767 (2010).
Bruce, M. C., Alano, P., Duthie, S. & Carter, R. Commitment of the malaria parasite Plasmodium falciparum to sexual and asexual development. Parasitology 100, 191–200 (1990).
Otto, T. D. et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 12, 86 (2014).
Mair, G. R. et al. Regulation of sexual development of Plasmodium by translational repression. Science 313, 667–669 (2006).
Lal, K. et al. Plasmodium male development gene-1 (mdv-1) is important for female sexual development and identifies a polarised plasma membrane during zygote development. Int. J. Parasitol. 39, 755–761 (2009).
Bushell, E. et al. Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272 (2017).
Yeoh, L. M., Goodman, C. D., Mollard, V., McFadden, G. I. & Ralph, S. A. Comparative transcriptomics of female and male gametocytes in Plasmodium berghei and the evolution of sex in alveolates. Genomics 18, 734 (2017).
Campbell, T. L., De Silva, E. K., Olszewski, K. L., Elemento, O. & Llinás, M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 6, e1001165 (2010).
Poran, A. et al. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature 551, 95–99 (2017).
Tibúrcio, M., Sauerwein, R., Lavazec, C. & Alano, P. Erythrocyte remodeling by Plasmodium falciparum gametocytes in the human host interplay. Trends Parasitol. 31, 270–278 (2015).
Painter, H. J., Carrasquilla, M. & Llinás, M. Capturing in vivo RNA transcriptional dynamics from the malaria parasite Plasmodium falciparum. Genome Res. 27, 1074–1086 (2017).
Lin, J. et al. A novel ‘gene insertion/marker out’ (GIMO) method for transgene expression and gene complementation in rodent malaria parasites. PLoS ONE 6, e29289 (2011).
Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15, 532-4–536-7 (1993).
Kozarewa, I. et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods 6, 291–295 (2009).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Modrzynska, K. et al. A knockout screen of ApiAP2 genes reveals networks of interacting transcriptional regulators controlling the Plasmodium life cycle. Cell Host Microbe 21, 11–22 (2017).
Guerreiro, A. et al. Genome-wide RIP-Chip analysis of translational repressor-bound mRNAs in the Plasmodium gametocyte. Genome Biol. 15, 493 (2014).
Young, J. A. et al. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol. Biochem. Parasitol. 143, 67–79 (2005).
Bailey, T. L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659 (2011).
Acknowledgements
We thank R. Menard and D. Bargieri for the diCre test plasmid used in this study, and M. Sanders and the WTSI sequencing service for assistance with RNA-seq sample processing. K.K.M. is supported by the Wellcome Trust and the Royal Society (ref. 202600/Z/16/Z). R.S.K. is supported by BBSRC (ref. BB/J013854/1). A.P.W. is supported by the Wellcome Trust (refs 083811 and 107046). O.B. is supported by the Wellcome Trust Sanger Institute (ref. WT098051).
Author information
Authors and Affiliations
Contributions
R.S.K. generated and phenotyped HP diCre, diCre test and PBANKA_0312700 lines, performed phenotyping experiments on the PBGAMi line and generated parasites for transcriptome sequencing. K.K.M. generated the AP2-G overexpression construct and PBGAMi line, performed phenotyping experiments on the PBGAMi line, generated RNA-seq libraries and performed RNA-seq data analysis. R.C. generated 820 diCre and GIMO lines and parasites for transcriptome sequencing. N.P. performed the ookinete conversion assay for PBANKA_0312700 line. O.B. and A.P.W. led and supervised the study. K.K.M. and A.P.W. wrote the manuscript with contributions from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Materials, Supplementary Figures 1–9, legends for Supplementary Tables 1–3 and original blots.
Supplementary Table 1
List of oligonucleotide primers used in the study.
Supplementary Table 2
Differential gene expression analysis between PBGAMiR+ and PBGAMiR populations at various time points post induction.
Supplementary Table 3
Gene ontology terms enriched in genes differentially expressed at different time points post induction.
Rights and permissions
About this article
Cite this article
Kent, R.S., Modrzynska, K.K., Cameron, R. et al. Inducible developmental reprogramming redefines commitment to sexual development in the malaria parasite Plasmodium berghei. Nat Microbiol 3, 1206–1213 (2018). https://doi.org/10.1038/s41564-018-0223-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0223-6
This article is cited by
-
Sexual differentiation in human malaria parasites is regulated by competition between phospholipid metabolism and histone methylation
Nature Microbiology (2023)
-
The transcriptional regulator HDP1 controls expansion of the inner membrane complex during early sexual differentiation of malaria parasites
Nature Microbiology (2022)
-
A transcriptional switch controls sex determination in Plasmodium falciparum
Nature (2022)
-
CRISPR/Cas9-engineered inducible gametocyte producer lines as a valuable tool for Plasmodium falciparum malaria transmission research
Nature Communications (2021)
-
Investigating a Plasmodium falciparum erythrocyte invasion phenotype switch at the whole transcriptome level
Scientific Reports (2020)