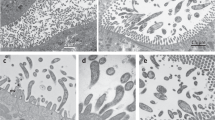
Abstract
Many anaerobic microbial parasites possess highly modified mitochondria known as mitochondrion-related organelles (MROs). The best-studied of these are the hydrogenosomes of Trichomonas vaginalis and Spironucleus salmonicida, which produce ATP anaerobically through substrate-level phosphorylation with concomitant hydrogen production; and the mitosomes of Giardia intestinalis, which are functionally reduced and lack any role in ATP production. However, to understand the metabolic specializations that these MROs underwent in adaptation to parasitism, data from their free-living relatives are needed. Here, we present a large-scale comparative transcriptomic study of MROs across a major eukaryotic group, Metamonada, examining lineage-specific gain and loss of metabolic functions in the MROs of Trichomonas, Giardia, Spironucleus and their free-living relatives. Our analyses uncover a complex history of ATP production machinery in diplomonads such as Giardia, and their closest relative, Dysnectes; and a correlation between the glycine cleavage machinery and lifestyles. Our data further suggest the existence of a previously undescribed biochemical class of MRO that generates hydrogen but is incapable of ATP synthesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Stairs, C. W., Leger, M. M. & Roger, A. J. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Phil. Trans. R. Soc. Lond. B 370, 20140326 (2015).
Lindmar, k, D. G. & Müller, M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J. Biol. Chem. 248, 7724–7728 (1973).
Müller, M. et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495 (2012).
Jerlstrom-Hultqvist, J. et al. Hydrogenosomes in the diplomonad Spironucleus salmonicida. Nat. Commun. 4, 2493 (2013).
Jedelsky, P. L. et al. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS ONE 6, e17285 (2011).
Karnkowska, A. et al. A eukaryote without a mitochondrial organelle. Curr. Biol. 26, 1274–1284 (2016).
Takishita, K. et al. Multigene phylogenies of diverse Carpediemonas-like organisms identify the closest relatives of ‘amitochondriate’ diplomonads and retortamonads. Protist 163, 344–355 (2012).
Simpson, A. G. & Patterson, D. J. On core jakobids and excavate taxa: the ultrastructure of Jakoba incarcerata. J. Eukaryot. Microbiol. 48, 480–492 (2001).
Dolezal, P., Likic, V., Tachezy, J. & Lithgow, T. Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318 (2006).
Martincova, E. et al. Probing the biology of Giardia intestinalis mitosomes using in vivo enzymatic tagging. Mol. Cell. Biol. 35, 2864–2874 (2015).
Neupert, W. & Herrmann, J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 (2007).
Yaffe, M. P., Ohta, S. & Schatz, G. A yeast mutant temperature-sensitive for mitochondrial assembly is deficient in a mitochondrial protease activity that cleaves imported precursor polypeptides. EMBO J. 4, 2069–2074 (1985).
Harris, S. R., Matus, A., Hrdy, I. & Kute , E. Reductive evolution of the mitochondrial processing peptidases of the unicellular parasites Trichomonas vaginalis and Giardia intestinalis. PLoS Pathog. 4, e1000243 (2008).
Garg, S. et al. Conservation of transit peptide-independent protein import into the mitochondrial and hydrogenosomal matrix. Genome Biol. Evol. 7, 2716–2726 (2015).
Kikuchi, G. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol. Cell. Biochem. 1, 169–187 (1973).
Mukherjee, M., Brown, M. T., McArthur, A. G. & Johnson, P. J. Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis. Eukaryot. Cell 5, 2062–2071 (2006).
Morrison, H. G. et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 317, 1921–1926 (2007).
Zubacova, Z. et al. The mitochondrion-like organelle of Trimastix pyriformis contains the complete glycine cleavage system. PLoS ONE 8, e55417 (2013).
Hampl, V. et al. Genetic evidence for a mitochondriate ancestry in the ‘amitochondriate’ flagellate Trimastix pyriformis. PLoS ONE 3, e1383 (2008).
Nývltová, E., Smutná, T., Tachezy, J. & Hrdý, I. OsmC and incomplete glycine decarboxylase complex mediate reductive detoxification of peroxides in hydrogenosomes of Trichomonas vaginalis. Mol. Biochem. Parasitol. 206, 29–38 (2016).
Xu, F. et al. On the reversibility of parasitism: adaptation to a free-living lifestyle via gene acquisitions in the diplomonad Trepomonas sp. PC1. BMC Biol. 14, 62 (2016).
Hampson, R. K., Barron, L. L. & Olson, M. S. Regulation of the glycine cleavage system in isolated rat liver mitochondria. J. Biol. Chem. 258, 2993–2999 (1983).
Steinbüchel, A. & Müller, M. Anaerobic pyruvate metabolism of Tritrichomonas foetus and Trichomonas vaginalis hydrogenosomes. Mol. Biochem. Parasitol. 20, 57–65 (1986).
Van Hellemond, J. J., Klockiewicz, M., Gaasenbeek, C. P., Roos, M. H. & Tielens, A. G. Rhodoquinone and complex II of the electron transport chain in anaerobically functioning eukaryotes. J. Biol. Chem. 270, 31065–31070 (1995).
Tielens, A. G. M., van Grinsven, K. W. A., Henze, K., van Hellemond, J. J. & Martin, W. Acetate formation in the energy metabolism of parasitic helminths and protists. Int. J. Parasitol. 40, 387–397 (2010).
Sanchez, L. B. & Müller, M. Purification and characterization of the acetate forming enzyme, acetyl-CoA synthetase (ADP-forming) from the amitochondriate protist, Giardia lamblia. FEBS Lett. 378, 240–244 (1996).
Noguchi, F. et al. Metabolic capacity of mitochondrion-related organelles in the free-living anaerobic stramenopile Cantina marsupialis. Protist 166, 534–550 (2015).
Field, J., Rosenthal, B. & Samuelson, J. Early lateral transfer of genes encoding malic enzyme, acetyl-CoA synthetase and alcohol dehydrogenases from anaerobic prokaryotes to Entamoeba histolytica. Mol. Microbiol. 38, 446–455 (2000).
Nývltová, E. et al. Lateral gene transfer and gene duplication played a key role in the evolution of Mastigamoeba balamuthi hydrogenosomes. Mol. Biol. Evol. 32, 1039–1055 (2015).
Schneider, R. E. et al. The Trichomonas vaginalis hydrogenosome proteome is highly reduced relative to mitochondria, yet complex compared with mitosomes. Int. J. Parasitol. 41, 1421–1434 (2011).
Murcha, M. W., Narsai, R., Devenish, J., Kubiszewski-Jakubiak, S. & Whelan, J. MPIC: a mitochondrial protein import components database for plant and non-plant species. Plant Cell Physiol. 56, e10 (2015).
Zhang, Q. et al. Marine isolates of Trimastix marina form a plesiomorphic deep-branching lineage within Preaxostyla, separate from other known Trimastigids (Paratrimastix n. gen.). Protist 166, 468–491 (2015).
Kolisko, M. et al. A wide diversity of previously undetected free-living relatives of diplomonads isolated from marine/saline habitats. Env. Microbiol. 12, 2700–2710 (2010).
Chevreux, B. et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14, 1147–1159 (2004).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Gentekaki, E. et al. Large-scale phylogenomic analysis reveals the phylogenetic position of the problematic taxon Protocruzia and unravels the deep phylogenetic affinities of the ciliate lineages. Mol. Phylogenet. Evol. 78, 36–42 (2014).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Criscuolo, A. & Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Minh, B. Q., Nguyen, M. A. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Lartillot, N., Rodrigue, N., Stubbs, D. & Richer, J. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615 (2013).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Capella-Gutierrez, S., Silla-Martinez, J. M. & Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Katoh, K. & Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Br. Bioinform. 9, 286–298 (2008).
Noguchi, F., Tanifuji, G., Brown, M. W., Fujikura, K. & Takishita, K. Complex evolution of two types of cardiolipin synthase in the eukaryotic lineage stramenopiles. Mol. Phylogenet. Evol. 101, 133–141 (2016).
Claros, M. G. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput. Appl. Biosci. 11, 441–447 (1995).
Emanuelsson, O., Brunak, S., von Heijne, G. & Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971 (2007).
Fukasawa, Y. et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics 14, 1113–1126 (2015).
Acknowledgements
M.K., M.M.L. and C.W.S. were supported by a grant (MOP-142349) from the Canadian Institutes of Health Research awarded to A.J.R. This work was also supported, in part, by a grant from the JSPS Strategic Young Researcher Overseas Visits Program (awarded to R.K.), by NSERC Grant 298366-2009 to A.G.B.S., by a Czech Science Foundation grant to I.Č. (project GA14-14105S) and by grants from the Japan Society for the Promotion of Science (JSPS; nos 15H05606 and 15K14591 awarded to R.K., 23117005 and 15H05231 awarded to T.H., and 23117006 awarded to Y.I.). We thank A. A. Heiss for his help with Trimastix marina data generation, and N. Ros for her comments on the manuscript.
Author information
Authors and Affiliations
Contributions
M.K., R.K., J.O.A., Y.I., A.G.B.S., T.H. and A.J.R. conceived and designed the experiments; M.K., K.K., I.Č., J.D.S., F.X., A.Y. and Q.Z. performed the experiments; M.M.L., M.K., R.K., C.W.S., K.K., L.E. and Y.I. analysed the data; R.K., C.W.S., J.D.S., K.T., Y.I., A.G.B.S., T.H. and A.J.R. contributed materials and/or analysis tools; and M.M.L., M.K., R.K., A.G.B.S. and A.J.R. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1–20, Supplementary Table 1 (PDF 1444 kb)
Supplementary Data 1
Predicted mitochondrion-related organelle proteins, and selected predicted cytosolic proteins, in metamonads. (XLSX 149 kb)
Supplementary Data 2
File containing the raw phylogenetic trees depicted in Supplementary Figs 4–20, including bootstrap support values, in Newick format. (TXT 539 kb)
Rights and permissions
About this article
Cite this article
Leger, M., Kolisko, M., Kamikawa, R. et al. Organelles that illuminate the origins of Trichomonas hydrogenosomes and Giardia mitosomes. Nat Ecol Evol 1, 0092 (2017). https://doi.org/10.1038/s41559-017-0092
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-017-0092
This article is cited by
-
The divergent ER-mitochondria encounter structures (ERMES) are conserved in parabasalids but lost in several anaerobic lineages with hydrogenosomes
BMC Biology (2023)
-
Mitochondrial metabolism of the facultative parasite Chilodonella uncinata (Alveolata, Ciliophora)
Parasites & Vectors (2023)
-
Complete mitochondrial genomes of Lycosa grahami and Lycosa sp. (Araneae: Lycosidae): comparison within the family Lycosidae
International Journal of Tropical Insect Science (2023)
-
Combined nanometric and phylogenetic analysis of unique endocytic compartments in Giardia lamblia sheds light on the evolution of endocytosis in Metamonada
BMC Biology (2022)
-
Anaerobic derivates of mitochondria and peroxisomes in the free-living amoeba Pelomyxa schiedti revealed by single-cell genomics
BMC Biology (2022)