Abstract
Endocytosis mediates the cellular uptake of micronutrients and the turnover of plasma membrane proteins. Clathrin-mediated endocytosis is the major uptake pathway in resting cells1, but several clathrin-independent endocytic routes exist in parallel2,3. One such pathway, fast endophilin-mediated endocytosis (FEME), is not constitutive but triggered upon activation of certain receptors, including the β1 adrenergic receptor4. FEME activates promptly following stimulation as endophilin is pre-enriched by the phosphatidylinositol-3,4-bisphosphate-binding protein lamellipodin4,5. However, in the absence of stimulation, endophilin foci abort and disassemble after a few seconds. Looking for additional proteins involved in FEME, we found that 20 out of 65 BAR domain-containing proteins tested colocalized with endophilin spots. Among them, FBP17 and CIP4 prime the membrane of resting cells for FEME by recruiting the 5′-lipid phosphatase SHIP2 and lamellipodin to mediate the local production of phosphatidylinositol-3,4-bisphosphate and endophilin pre-enrichment. Membrane-bound GTP-loaded Cdc42 recruits FBP17 and CIP4, before being locally deactivated by RICH1 and SH3BP1 GTPase-activating proteins. This generates the transient assembly and disassembly of endophilin spots, which lasts 5–10 seconds. This mechanism periodically primes patches of the membrane for prompt responses upon FEME activation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
06 August 2018
In the version of this Letter originally published, on the x axis of the right panel of Fig. 2h the labels ‘+Dobu’ and ‘+Pi3Ki’ were slightly misplaced, which meant the label ‘ClP4OEx’ was partly obscured. This has now been amended in all online versions of the Letter.
20 August 2018
In the version of this Letter originally published, the name of co-author Safa Lucken-Ardjomande Häsler was coded wrongly, resulting in it being incorrect when exported to citation databases. This has been corrected, though no visible changes will be apparent.
References
Bitsikas, V., Correa, I. R. Jr & Nichols, B. J. Clathrin-independent pathways do not contribute significantly to endocytic flux. eLife 3, e03970 (2014).
Johannes, L., Parton, R. G., Bassereau, P. & Mayor, S. Building endocytic pits without clathrin. Nat. Rev. Mol. Cell Biol. 16, 311–321 (2015).
Ferreira, A. P. A. & Boucrot, E. Mechanisms of carrier formation during clathrin-independent endocytosis. Trends Cell Biol. 28, 188–200 (2018).
Boucrot, E. et al. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 517, 460–465 (2015).
Vehlow, A. et al. Endophilin, lamellipodin, and mena cooperate to regulate F-actin-dependent EGF-receptor endocytosis. EMBO J. 32, 2722–2734 (2013).
Watanabe, S. & Boucrot, E. Fast and ultrafast endocytosis. Curr. Opin. Cell Biol. 47, 64–71 (2017).
Ehrlich, M. et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 118, 591–605 (2004).
Cestra, G. et al. The SH3 domains of endophilin and amphiphysin bind to the proline-rich region of synaptojanin 1 at distinct sites that display an unconventional binding specificity. J. Biol. Chem. 274, 32001–32007 (1999).
Nakano-Kobayashi, A., Kasri, N. N., Newey, S. E. & Van Aelst, L. The Rho-linked mental retardation protein OPHN1 controls synaptic vesicle endocytosis via endophilin A1. Curr. Biol. 19, 1133–1139 (2009).
Sanchez-Barrena, M. J. et al. Bin2 is a membrane sculpting N-BAR protein that influences leucocyte podosomes, motility and phagocytosis. PLoS ONE 7, e52401 (2012).
Frost, A., Unger, V. M. & De Camilli, P. The BAR domain superfamily: membrane-molding macromolecules. Cell 137, 191–196 (2009).
Rao, Y. & Haucke, V. Membrane shaping by the Bin/amphiphysin/Rvs (BAR) domain protein superfamily. Cell Mol. Life Sci. 68, 3983–3993 (2011).
Nishimura, T., Morone, N. & Suetsugu, S. Membrane re-modelling by BAR domain superfamily proteins via molecular and non-molecular factors. Biochem. Soc. Trans. 46, 379–389 (2018).
Eichel, K., Jullie, D. & von Zastrow, M. β-Arrestin drives MAP kinase signalling from clathrin-coated structures after GPCR dissociation. Nat. Cell Biol. 18, 303–310 (2016).
Itoh, T. et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell 9, 791–804 (2005).
Shimada, A. et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129, 761–772 (2007).
Taylor, M. J., Perrais, D. & Merrifield, C. J. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9, e1000604 (2011).
Tsujita, K., Takenawa, T. & Itoh, T. Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat. Cell Biol. 17, 749–758 (2015).
Xiong, D. et al. Frequency and amplitude control of cortical oscillations by phosphoinositide waves. Nat. Chem. Biol. 12, 159–166 (2016).
Pesesse, X. et al. The Src homology 2 domain containing inositol 5-phosphatase SHIP2 is recruited to the epidermal growth factor (EGF) receptor and dephosphorylates phosphatidylinositol 3,4,5-trisphosphate in EGF-stimulated COS-7 cells. J. Biol. Chem. 276, 28348–28355 (2001).
Aspenstrom, P. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 7, 479–487 (1997).
Ho, H. Y. et al. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP–WIP complex. Cell 118, 203–216 (2004).
Billuart, P. et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature 392, 923–926 (1998).
Richnau, N. & Aspenstrom, P. Rich, a rho GTPase-activating protein domain-containing protein involved in signaling by Cdc42 and Rac1. J. Biol. Chem. 276, 35060–35070 (2001).
Elbediwy, A. et al. Epithelial junction formation requires confinement of Cdc42 activity by a novel SH3BP1 complex. J. Cell Biol. 198, 677–693 (2012).
Bertot, L. et al. Quantitative and statistical study of the dynamics of clathrin-dependent and -independent endocytosis reveal a differential role of endophilinA2. Cell Rep. 22, 1574–1588 (2018).
Renard, H. F. et al. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature 517, 493–496 (2015).
Sabharanjak, S., Sharma, P., Parton, R. G. & Mayor, S. GPI-anchored proteins are delivered to recycling endosomes via a distinct Cdc42-regulated, clathrin-independent pinocytic pathway. Dev. Cell 2, 411–423 (2002).
Lundmark, R. et al. The GTPase-activating protein GRAF1 regulates the CLIC/GEEC endocytic pathway. Curr. Biol. 18, 1802–1808 (2008).
Howes, M. T. et al. Clathrin-independent carriers form a high capacity endocytic sorting system at the leading edge of migrating cells. J. Cell Biol. 190, 675–691 (2010).
Sathe, M. et al. Small GTPases and BAR domain proteins regulate branched actin polymerisation for clathrin and dynamin-independent endocytosis. Nat. Commun. 9, 1835 (2018).
Francis, M. K. et al. Endocytic membrane turnover at the leading edge is driven by a transient interaction between Cdc42 and GRAF1. J. Cell Sci. 128, 4183–4195 (2015).
Henne, W. M. et al. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science 328, 1281–1284 (2010).
Frost, A. et al. Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817 (2008).
Carmona, G. et al. Lamellipodin promotes invasive 3D cancer cell migration via regulated interactions with Ena/VASP and SCAR/WAVE. Oncogene 35, 5155–5169 (2016).
Hoekstra, E. et al. Lipid phosphatase SHIP2 functions as oncogene in colorectal cancer by regulating PKB activation. Oncotarget 7, 73525–73540 (2016).
Tao, Y. et al. SH3-domain binding protein 1 in the tumor microenvironment promotes hepatocellular carcinoma metastasis through WAVE2 pathway. Oncotarget 7, 18356–18370 (2016).
Baldassarre, T. et al. Endophilin A2 promotes TNBC cell invasion and tumor metastasis. Mol. Cancer Res 13, 1044–1055 (2015).
Rolland, Y. et al. The CDC42-interacting protein 4 controls epithelial cell cohesion and tumor dissemination. Dev. Cell 30, 553–568 (2014).
Endris, V. et al. SrGAP3 interacts with lamellipodin at the cell membrane and regulates Rac-dependent cellular protrusions. J. Cell Sci. 124, 3941–3955 (2011).
Krause, M. et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell 7, 571–583 (2004).
Doherty, G. J. et al. The endocytic protein GRAF1 is directed to cell–matrix adhesion sites and regulates cell spreading. Mol. Biol. Cell 22, 4380–4389 (2011).
Vizcaíno, J. A. et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 44, D447–D456 (2016).
Acknowledgements
We thank S. Y. Peak-Chew for mass spectrometry, M. Edwards, M. Dumoux and K. McGourty for technical help, P. De Camilli (Yale University), P. Aspenstrom (Karolinska Institute), P. Randazzo (NIH), M. Negishi (Kyoto University), G. Rappold (University of Heidelberg), H. Kent (MRC Laboratory of Molecular Biology), J. Gallop (University of Cambridge), T. Takeda (MRC Molecular Biology), M. Parsons (King’s College London), F. Gertler (MIT) and T. Kirchhausen (Harvard Medical School) for the kind gift of reagents and the members of the Boucrot lab for helpful comments. E.B. was a Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips Research Fellow (BB/R01551X/1), a Lister Institute Research Fellow and a recipient of a grant from the Royal Society Research Grant (RG120481). S.L.-A.H. and H.T.M. were supported by the MRC UK (grant number U105178805 to H.T.M.) and by the Swiss National Science Foundation (fellowship number PA00P3-124164 to S.L.-A.H). A.P.A.F was supported by the Fundação para a Ciência e Tecnologia.
Author information
Authors and Affiliations
Contributions
E.B. designed the research and supervised the project. L.C.W.H., S.K., I.D.M. and E.B. performed the cell biology experiments. L.C.W.H., S.K., A.-L.L., A.P.A.F., E.B. and H.T.M. performed the pull-down experiments. S.L.-A.H., I.D.M., S.K. and L.M.Q. provided critical reagents. M.K. helped to design and supervise some of the experiments. E.B. wrote the manuscript with input from all of the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
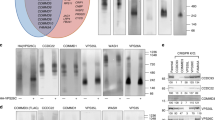
Supplementary Figure 1 BAR domain proteins binding to Endophilin.
a, Pull down using GST-SH3 domains for Endophilin A1, A2 or A3 and rat brain lysate. Interacting proteins were identified by mass spectrometry. Known interactors are shown in grey, members of the WAVE complex in black and BAR-domain proteins in blue. Asterisk denotes GST-SH3. List of identified proteins is provided in Supplementary Table 1. b, Pull-down using GST-SH3 domains of Endophilin A2 and cell extracts expressing EGFP-tagged indicated BAR proteins. Bin2 and OPHN1 were used as positive controls. GST was used as negative control. Binding proteins were detected by immunoblotting with an anti-EGFP antibody. ‘input’ lanes correspond to 1-5 % of the cell extracts. c, Schematic depicting the domain organization of the human full-length BAR domain-containing proteins cloned and used in this study. d, co-immunoprecipitation assays from cells co-expressing the indicated combination of EGFP- or Myc-tagged BAR domains from FBP17, CIP4 and TOCA-1 and showing that all three can heterodimerize. Input (‘I’, which corresponds to 10% of cell extracts), unbound (‘U’) and bound (‘B’) fractions for each were immunoblotted with anti-EGFP and anti-Myc antibodies. All experiments were repeated independently at least three times with similar results. Unprocessed original scans are provided in Supplementary Fig. 6.
Supplementary Figure 2 BAR domain-containing proteins tissue expression pattern.
a, Number of tissues where expressed sequence tag (EST) were detected in the UniGene transcriptome database for the named human BAR domain-containing proteins. Zones for expression in 25% and 50% of the tissues are highlighted in dark and light red, respectively. b, Number of transcript per million (TPM) in kidney (tissue of origin of BSC1 cells) for the named human BAR domain-containing proteins recorded in the UniGene transcriptome database. Red boxes highlights genes for which no transcript were detected. c, Number of transcript per million (TPM) in eye (tissue of origin of RPE1 cells) for the named human BAR domain-containing proteins recorded in the UniGene transcriptome database. Red boxes highlights genes for which no transcript were detected. The data were recorded from the NCBI UniGene transcriptome database (see Methods for details).
Supplementary Figure 3 BAR proteins colocalizing with Endophilin in resting cells.
a, Confocal microscopy sections showing an example of the named BAR proteins tagged with EGFP at their C-termini colocalizing (RICH1-EGFP, left) or not (RICH2-EGFP, right) with endogenous Endophilin in BSC1 cells. b, Criteria used to score BAR candidates negative or positive. Intensity profiles were acquired along the indicated lines. c-f, Confocal microscopy sections showing the colocalization of the named controls (soluble EGFP and CAAX-EGFP), BAR, N-BAR or BAR-PH proteins tagged with EGFP at their C-termini and Endophilin. Images were cropped from larger pictures and angled as in (a) so that all images are oriented similarly. Arrowheads point to Endophilin spots colocalizing with BAR proteins at the cell surface. Images are representative of 10 captures from three independent biological experiments. All experiments were repeated independently at least three times with similar results. Scale bars, 5 μm.
Supplementary Figure 4 BAR proteins colocalizing with Endophilin in resting cells (continued).
a-c, Confocal microscopy sections showing the colocalization of the named PX-BAR, F-BAR or I-BAR proteins tagged with EGFP at their C-termini and Endophilin in BSC1 cells. Arrowheads point to Endophilin spots colocalizing with BAR proteins at the cell surface. Images are representative of 10 captures from three independent biological experiments. d, Absence of colocalization between internalized Alexa488-Transferrin and endogenous Endophilin in BSC1 and RPE1 cells stimulated with 10μM dobutamine and 50μg/mL Alexa488-Transferrin for 30sec at 37 °C for 30sec. Images are representative of 10 captures from three independent biological experiments. e, Top, β1-AR and GAPDH levels in the indicated plasma membrane and total fractions from BSC1 cells depleted or not for Endophilin (Endo TKD), AP2, FBP17 and CIP4 (F+C DKD) or Amphiphysin and Bin1 (A+B DKD). Unprocessed original scans are provided in Supplementary Fig. 6. f, Colocalization of β1-adrenergic receptor (β1-AR) and LAMP1 in resting BSC1 cells or upon incubation with 10μM dobutamine for 30min. Images are representative of 6 captures from three independent biological experiments. All experiments were repeated independently at least three times with similar results. Scale bars, 20 (f) and 5μm (a,,b,c,d).
Supplementary Figure 5 Cdc42 is deactivated locally by RICH1 and SH3BP1.
a, Confocal microscopy sections showing the transient increase in endogenous CIP4 and Endophilin on FEME carriers after 2 but not 10min incubation with 10μM ML141 (‘Cdc42i 1’). Images are representative of 6 captures from three independent biological experiments. b, Confocal microscopy section showing the decrease or not in endogenous Endophilin recruitment at the leading edge of resting cells overexpressing wild-type EGFP-tagged RICH2, SH3BP1 or OPHN1 but not their R291A, R312A and R409A respective GAP mutants (GAP*). Arrowheads point to Endophilin spots at the cell surface. Images are representative of 6 captures from three independent biological experiments. c,d, Recruitment of endogenous RICH1, Lpd and CIP4 in resting BSC1 cells depleted or not for Lamellipodin (Lpd KD), RICH1 and SH3BP1 (R+S DKD) or FBP17 and CIP4 (F+C DKD). Images are representative of 10 captures from three independent biological experiments. e, Pull-down using GST or GST-SH3 domains of Endophilin A2 or FBP17 and cell extracts expressing RICH1, RICH2, SH3BP1 or OPHN1 tagged with EGFP at their C-termini. Inputs correspond to 1 to 5% of the cell extracts. All experiments were repeated independently at least three times with similar results. Unprocessed original scans are provided in Supplementary Fig. 6. Bars, 5 μm.
Supplementary Figure 6
Unprocessed original scans of all gels and blots.
Supplementary information
Supplementary Information
Supplementary Figures 1–6, Supplementary Table and Supplementary Video legends.
Supplementary Table 1
List of interactors identified in Supplementary Fig. 1a.
Supplementary Table 2
Statistics Source Data.
Supplementary Table 3
Sequences of DNA and siRNA primers used in this study.
Supplementary Table 4
Information on antibodies used in this study.
Supplementary Video 1
Spinning-disk confocal microscopy of a resting BSC1 cell transiently expressing low levels of EGFP–LCa (clathrin, green) and endophilin A2-RFP (red) and imaged at 0.5 Hz.
Supplementary Video 2
Spinning-disk confocal microscopy of a resting BSC1 cell transiently expressing low levels of FBP17–EGFP (green) and Endophilin A2–RFP (red) and imaged at 0.5 Hz.
Supplementary Video 3
Spinning-disk confocal microscopy of a resting BSC1 cell transiently expressing low levels of CIP4–EGFP (green) and Endophilin A2–RFP (red) and imaged at 2 Hz. 5 nM GDC-0941 (PI3Ki) was added at timeframe 100.
Supplementary Video 4
Spinning-disk confocal microscopy of a resting BSC1 cell transiently expressing low levels of FBP17–EGFP (green) and Endophilin A2–RFP (red) and imaged at 2 Hz.
Supplementary Video 5
Spinning-disk confocal microscopy of a resting BSC1 cell transiently expressing low levels of CIP4–EGFP (green) and Endophilin A2–RFP (red) and imaged at 2 Hz.
Supplementary Video 6
Spinning-disk confocal microscopy of a resting BSC1 cell transiently expressing low levels of CIP4–EGFP (green) and Endophilin A2–RFP (red) and imaged at 2 Hz. 5 μM AS19499490 (‘SHIP2i’) was added at timeframe 100 and again at timeframe 200.
Rights and permissions
About this article
Cite this article
Chan Wah Hak, L., Khan, S., Di Meglio, I. et al. FBP17 and CIP4 recruit SHIP2 and lamellipodin to prime the plasma membrane for fast endophilin-mediated endocytosis. Nat Cell Biol 20, 1023–1031 (2018). https://doi.org/10.1038/s41556-018-0146-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0146-8
This article is cited by
-
Phosphoinositides as membrane organizers
Nature Reviews Molecular Cell Biology (2022)
-
Multivalent interactions between molecular components involved in fast endophilin mediated endocytosis drive protein phase separation
Nature Communications (2022)
-
Homeostatic membrane tension constrains cancer cell dissemination by counteracting BAR protein assembly
Nature Communications (2021)
-
Tutorial: using nanoneedles for intracellular delivery
Nature Protocols (2021)
-
Cdk5 and GSK3β inhibit fast endophilin-mediated endocytosis
Nature Communications (2021)