Abstract
Maintenance of endoplasmic reticulum (ER) proteostasis is controlled by a signalling network known as the unfolded protein response (UPR). Here, we identified filamin A as a major binding partner of the ER stress transducer IRE1α. Filamin A is an actin crosslinking factor involved in cytoskeleton remodelling. We show that IRE1α controls actin cytoskeleton dynamics and affects cell migration upstream of filamin A. The regulation of cytoskeleton dynamics by IRE1α is independent of its canonical role as a UPR mediator, serving instead as a scaffold that recruits and regulates filamin A. Targeting IRE1α expression in mice affected normal brain development, generating a phenotype resembling periventricular heterotopia, a disease linked to the loss of function of filamin A. IRE1α also modulated cell movement and cytoskeleton dynamics in fly and zebrafish models. This study unveils an unanticipated biological function of IRE1α in cell migration, whereby filamin A operates as an interphase between the UPR and the actin cytoskeleton.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
08 August 2018
In the version of this Article originally published, the competing interests statement was missing. The authors declare no competing interests; this statement has now been added in all online versions of the Article.
27 April 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41556-021-00673-2
References
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Hetz, C., Chevet, E. & Oakes, S. A. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 17, 829–838 (2015).
Urra, H., Dufey, E., Lisbona, F. & Rojas-Rivera, D. When ER stress reaches a dead end. Biochim. Biophys. Acta 1833, 3507–3517 (2013).
Oakes, S. A. & Papa, F. R. The role of endoplasmic reticulum stress in human pathology. Annu. Rev. Pathol. 10, 173–194 (2015).
Wang, M. & Kaufman, R. J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 (2016).
Calfon, M. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002).
Lee, K. et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16, 452–466 (2002).
Hetz, C. & Glimcher, L. H. Fine-tuning of the unfolded protein response: Assembling the IRE1alpha interactome. Mol. Cell 35, 551–561 (2009).
Feng, Y. & Walsh, C. A. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 6, 1034–1038 (2004).
Oono, K. et al. JAB1 participates in unfolded protein responses by association and dissociation with IRE1. Neurochem. Int. 45, 765–772 (2004).
Nakamura, F., Stossel, T. P. & Hartwig, J. H. The filamins: organizers of cell structure and function. Cell Adhes. Migr. 5, 160–169 (2011).
Sepulveda, D. et al. Interactome screening identifies the ER luminal chaperone Hsp47 as a regulator of the unfolded protein response transducer IRE1alpha. Mol. Cell 69, 238–252 (2018).
Nakamura, F., Osborn, T. M., Hartemink, C. A., Hartwig, J. H. & Stossel, T. P. Structural basis of filamin A functions. J. Cell Biol. 179, 1011–1025 (2007).
Riedl, J. et al. Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 (2008).
Ridley, A. J. Rho GTPases and cell migration. J. Cell Sci. 114, 2713–2722 (2001).
Vadlamudi, R. K. et al. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat. Cell Biol. 4, 681–690 (2002).
Baldassarre, M. et al. Filamins regulate cell spreading and initiation of cell migration. PLoS ONE 4, e7830 (2009).
Lynch, C. D. et al. Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol. Biol. Cell 22, 1263–1273 (2011).
van Vliet, A. R. et al. The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with filamin-A and F-actin remodeling. Mol. Cell 65, 885–899 (2017).
Ali, M. M. et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 30, 894–905 (2011).
Bouchecareilh, M., Higa, A., Fribourg, S., Moenner, M. & Chevet, E. Peptides derived from the bifunctional kinase/RNase enzyme IRE1α modulate IRE1α activity and protect cells from endoplasmic reticulum stress. FASEB J. 25, 3115–3129 (2011).
Zhou, A.-X., Hartwig, J. H. & Akyürek, L. M. Filamins in cell signaling, transcription and organ development. Trends Cell Biol. 20, 113–123 (2010).
Zhong, Z. et al. Cyclin D1/cyclin-dependent kinase 4 interacts with filamin A and affects the migration and invasion potential of breast cancer cells. Cancer Res. 70, 2105–2114 (2010).
Tigges, U., Koch, B., Wissing, J., Jockusch, B. M. & Ziegler, W. H. The F-actin cross-linking and focal adhesion protein filamin A is a ligand and in vivo substrate for protein kinase C alpha. J. Biol. Chem. 278, 23561–23569 (2003).
Sarkisian, M. R. et al. MEKK4 signaling regulates filamin expression and neuronal migration. Neuron 52, 789–801 (2006).
Xue, Z. et al. A conserved structural determinant located at the interdomain region of mammalian inositol-requiring enzyme 1α. J. Biol. Chem. 286, 30859–30866 (2011).
Li, H., Korennykh, A. V., Behrman, S. L. & Walter, P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc. Natl Acad. Sci. USA 107, 16113–16118 (2010).
Wood, W. & Jacinto, A. Drosophila melanogaster embryonic haemocytes: masters of multitasking. Nat. Rev. Mol. Cell Biol. 8, 542–551 (2007).
Lee, T. & Luo, L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254 (2001).
Solnica-Krezel, L. & Sepich, D. S. Gastrulation: making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 28, 687–717 (2012).
Auf, G. et al. Inositol-requiring enzyme 1α is a key regulator of angiogenesis and invasion in malignant glioma. Proc. Natl Acad. Sci. USA 107, 15553–15558 (2010).
Nguyen, D. T. et al. Nck-dependent activation of extracellular signal-regulated kinase-1 and regulation of cell survival during endoplasmic reticulum stress. Mol. Biol. Cell 15, 4248–4260 (2004).
Hammerschmidt, M. et al. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development 123, 143–151 (1996).
Waxman, J. S., Hocking, A. M., Stoick, C. L. & Moon, R. T. Zebrafish Dapper1 and Dapper2 play distinct roles in Wnt-mediated developmental processes. Development 131, 5909–5921 (2004).
Behrndt, M. et al. Forces driving epithelial spreading in zebrafish gastrulation. Science 338, 257–260 (2012).
Fox, J. W. et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21, 1315–1325 (1998).
Sheen, V. L. et al. Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum. Mol. Genet. 10, 1775–1783 (2001).
Iwawaki, T., Akai, R., Yamanaka, S. & Kohno, K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc. Natl Acad. Sci. USA 106, 16657–16662 (2009).
Sarkisian, M. R., Bartley, C. M. & Rakic, P. Trouble making the first move: interpreting arrested neuronal migration in the cerebral cortex. Trends Neurosci. 31, 54–61 (2008).
Martinez, G., Khatiwada, S., Costa-Mattioli, M. & Hetz, C. ER proteostasis control of neuronal physiology and synaptic function. Trends Neurosci. https://doi.org/10.1016/j.tins.2018.05.009 (2018).
Laguesse, S. et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell 35, 553–567 (2015).
Savoy, R. M. & Ghosh, P. M. The dual role of filamin A in cancer: can’t live with (too much of) it, can’t live without it. Endocr. Relat. Cancer 20, R341–R356 (2013).
Hetz, C. et al. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science 312, 572–576 (2006).
Matsuda, T. & Cepko, C. L. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl Acad. Sci. USA 101, 16–22 (2004).
Lisbona, F. et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol. Cell 33, 679–691 (2009).
Rodriguez, D. A. et al. BH3-only proteins are part of a regulatory network that control the sustained signalling of the unfolded protein response sensor IRE1alpha. EMBO J. 31, 2322–2335 (2012).
Henriquez, D. R., Bodaleo, F. J., Montenegro-Venegas, C. & Gonzalez-Billault, C. The light chain 1 subunit of the microtubule-associated protein 1B (MAP1B) is responsible for Tiam1 binding and Rac1 activation in neuronal cells. PLoS ONE 7, e53123 (2012).
Wieckowski, M. R., Giorgi, C., Lebiedzinska, M., Duszynski, J. & Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 (2009).
Ramirez, O., Garcia, A., Rojas, R., Couve, A. & Hartel, S. Confined displacement algorithm determines true and random colocalization in fluorescence microscopy. J. Microsc. 239, 173–183 (2010).
Hetz, C. et al. The proapoptotic BCL-2 family member BIM mediates motoneuron loss in a model of amyotrophic lateral sclerosis. Cell Death Differ. 14, 1386–1389 (2007).
Urra, H. et al. Caveolin-1-enhanced motility and focal adhesion turnover require tyrosine-14 but not accumulation to the rear in metastatic cancer cells. PLoS ONE 7, e33085 (2012).
Lim, K. B. et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 283, 20454–20472 (2008).
Grande-Garcia, A. et al. Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J. Cell Biol. 177, 683–694 (2007).
Fuentes, P., Canovas, J., Berndt, F. A., Noctor, S. C. & Kukuljan, M. CoREST/LSD1 control the development of pyramidal cortical neurons. Cereb. Cortex 22, 1431–1441 (2012).
LoTurco, J., Manent, J. B. Sidiqi, F. New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb. Cortex 19, 20–25 (2009).
Moreira, C. G., Regan, J. C., Zaidman-Remy, A., Jacinto, A. & Prag, S. Drosophila hemocyte migration: an in vivo assay for directional cell migration. Methods Mol. Biol. 769, 249–260 (2011).
Barry, D.J., Durkin, C.H., Abella, J.V. & Way, M. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J. Cell Biol. 209, 163–180 (2015).
Barth, K. A. & Wilson, S. W. Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development 121, 1755–1768 (1995).
Yeh, C. -M. et al. Ptenb mediates gastrulation cell movements via Cdc42/AKT1 in zebrafish. PLoS ONE 6, e18702 (2011).
Acknowledgements
We thank P. Walter for providing IRE1-3F6H-GFP cells and M. P. Sheetz for providing filamin A-deficient cells and expression vectors. In addition, we thank D. Calderwood and D. Iwamoto for providing IgG-like filamin A individual plasmids tagged with GFP or GST. We thank L. Leyton for providing phospho-specific anti-PKC antibodies. This work was funded by the following bodies: FONDECYT no. 3160461 (to H.U.), no. 1140549 and 1180993 (to C.H.), no. 1140325 (to C.G.-B.), no. 1150608 (to R.L.V.); no. 1150766 (to F.C.); no. 3160478 (to E.P.); no. 3150113 (to A.C-S.) and no. 1140522 (to Á.G.); Millennium Institute No. P09-015-F (to C.H., A.C. and M.L.C.); FONDAP 15150012 (to C.H., F.C., C.G.-B. and M.L.C.); FONDAP 15090007 (to Á.G.); ECOS-CONICYT 170032 (to C.H.); PIA-CONICYT ACT1401 (to Á.G.); NIH R01 Gm113188 (L.Q.)and CONICYT ACT1402 (to M.L.C.). We also thank the following organizations: the European Commission R&D MSCA-RISE #734749; The Michael J Fox Foundation for Parkinson’s Research—Target Validation grant No 9277; FONDEF ID16I10223; FONDEF D11E1007; the US Office of Naval Research-Global (ONR-G) N62909-16-1-2003; the US Air Force Office of Scientific Research FA9550-16-1-0384; ALSRP Therapeutic Idea Award AL150111; Muscular Dystrophy Association 382453; and CONICYT-Brazil 441921/2016-7 (to C.H.). T.I. is supported by the Toray Science Foundation. J.C., C.M.L (no. 21160967) and D.R.H. are doctoral fellows supported by a CONICYT fellowship and by a CONICYT research grant.
Author information
Authors and Affiliations
Contributions
H.U and C.H. designed the study. H.U., D.R.H., J.C., D.V.-C., A.C.-S., E.C., E.P., E.M., Y.M.H., C.M.L., S.A.-R., R.F., R.L.V., R.A., D.A.R. and C.A:R. participated in experimental designs, performed experiments and analysed the data. R.L.V., F.C., A.C., L.Q., E.C., T.I., M.L.C., Á.G. and C.G.-B. supervised the experiments and participated in the designs. H.U and C.H. wrote or contributed to writing the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Yeast-two hybrid ontology analysis and control experiments.
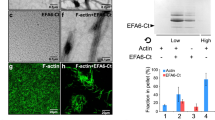
(A) Gene ontology association indicating the most representative molecular functions founded using Toppgene analysis top 10 strongest IRE1α interactors found in the yeast-two hybrid screening. The threshold significance is shown. (B) Cluster and functional analysis of top 10 strongest IRE1α interactors. Interaction found by Y2H screen is color coded by function and known interaction is indicated. Generation of IRE1α reconstituted and control cells: (C) IRE1α KO MEFs were stably transduced with retroviruses expressing the HA-tagged version of full-length human IRE1α (IRE1α-HA). The levels of IRE1α were evaluated by western blot using specific antibodies. Cells were treated with 100 ng ml–1 of Tm for 16 h and then Xbp1 mRNA splicing was evaluated by RT-PCR. PCR fragments corresponding to the Xbp1u or Xbp1s forms of XBP1 mRNA are indicated. The levels of Edem1, a transcriptional target of XBP1s and Blos1, a target of RIDD were analyzed by real-time PCR (n=2 independent experiments). Control experiment for co-localization: (D) Cell were transfected with plasmid coding for GFP and stained for actin using Phalloidin-Rodamine dye. Images are shown to illustrate the difference in the fluorescence localization of both staining. In all panels, data represents mean ± s.e.m. of indicated number of independent experiments.
Supplementary Figure 2 IRE1α and Filamin A regulate actin cytoskeleton dynamics.
(A) (Left panel) Raw data of each frame of the quantified green areas of Fig. 2a. (Right panel) Raw data of each frame of the quantified red areas of Fig. 2a. (B) Distribution of life-act across the cell was quantified. 0 and 1 represent the distribution of life-act fluorescence intensity from the cell periphery to the center of the cell, respectively (5 (WT), 5 (IRE1α KO) and 7 (IRE1α-HA) independent set of videos). (C) WT or FLNA KO cells transiently transfected with a plasmid encoding Life-Act were plated onto fibronectin-coated plates and recorded by time-lapse confocal microscopy every 30 s for 5 min. The average signal over time of the protruding and retracting cell area is shown. Protrusion-retraction velocity and filopodia number per cell calculated using ADAPT software are shown (8 (WT) and 7 (FLNA KO) independent set of videos). In all panels, data represents mean ± s.e.m. of indicated number of independent experiments; two-tailed t test was used, otherwise is indicated; *p < 0.05 and **p < 0.01.
Supplementary Figure 3 IRE1α regulate cell migration independent of PERK signaling.
IRE1α deficiency does not affect cell proliferation: (A) Cell counting of IRE1α KO MEFs expressing IRE1α-HA or empty vector (Mock) were stimulated with FSB (n=3 independent experiments). Filamin A regulates cell migration: (B) Western blot of total protein extracts of WT or Filamin A KO MEFs. Right panel: wound-healing assay of WT or Filamin A KO was performed (n=3 independent experiments). IRE1α expression in multiple cell types transfected with siRNAs: (C) Indicated cell lines were transfected with siRNAs for IRE1α or controls for 48 h. The levels of IRE1α and actin mRNA were analyzed by real-time PCR. Control experiments: (D) Left panel: western blot of total protein extracts of cells transfected with plasmids coding for IRE1α-HA or empty. Right panel: Boyden chamber assay of transfected cells (two-tailed t test, n=4 independent experiments). Control experiments for cell transfection efficiency: (E) flow cytometry of MEFs expressing expression vectors for Luciferase (shLuc) or IRE1α (shIRE1α) were transiently transfected with pEGFP in addition to Filamin A WT (FLNA-MYC) (n=2 independent experiments). (F) Flow cytometry of WT or FLNA KO MEFs transiently transfected with pEGFP in addition to IRE1α-HA expressing plasmids (n=2 independent experiments). Effects of PERK on cell migration: (G) Left panel: western blot of total proteins extracts of IRE1α KO MEFs mock or IRE1α-HA transfected with control or PERK siRNAs. Right panel: Boyden chamber assay of transfected cells was assessed (n=4 independent experiments). (H) Boyden chamber assay of MEF cells pretreated for 2 h with 50 µM of PERK inhibitor GSK2606414 (n=4 independent experiments). (I) Left panel: Confirmation of knockdown efficiency using qPCR (n=2 independent experiments). Right panel: Boyden chamber assay of shLuc, shIRE1α and shATF6 cells was performed (n=3 independent experiments). In all panels, data represents mean ± s.e.m. of indicated number of independent experiments; one-way ANOVA followed by Tukey’s test was used, otherwise is indicated; *p < 0.05 and **p < 0.01.
Supplementary Figure 4 The C-terminal distal region on IRE1α is involved in actin cytoskeleton dynamics.
IRE1α KO MEFs stably transduced with retroviral expression vectors for IRE1α-HA WT or the Δ965 mutant and then were transiently transfected with a plasmid encoding Life-Act. Cells were plated in fibronectin-coated plates and recorded by time-lapse confocal microscopy every 30 s for 5 min. Segmented images are shown. Additional quantifications are presented in Fig. 4h (20 (IRE1α WT) and 14 (IRE1αΔ965) independent set of videos). In all panels, data represents mean ± s.e.m. of indicated number of independent experiments; two-tailed t test was used, otherwise is indicated; *p < 0.05 and **p < 0.01.
Supplementary Figure 5 IRE1α regulate Filamin A phosphorylation at serine 2152 through PKCα.
Contribution of PERK pathways to Filamin A phosphorylation: (A) western blot of total protein extracts to evaluate Filamin A phosphorylation of IRE1α KO MEFs transfected with control or PERK siRNAs and treated with FBS or pretreated with GSK2606414. IRE1α regulates cytoskeleton dynamics upstream of Filamin A: (B) Time-lapse confocal microscopy IRE1α KO MEFs transiently transfected Life-Act in addition to a Filamin A WT (FLNA-MYC) or S2152A mutant vectors. Segmented images are shown. (C) Filopodia number per cell calculated using ADAPT software (5 (IRE1α WT), 11 (IRE1α-KO), 7 (IRE1α-KO + Filamin A WT) and 7 (IRE1α-KO + Filamin A S2152A) independent sets of videos). (D) Average signal over time of the protruding and retracting cell area of cells from Fig. 5f was quantified. Control experiments: (E) western blot of total protein extracts of MEFs cells expressing shLuc or shIRE1α transiently transfected with pEGFP in addition to Filamin A WT (FLNA-MYC) or S2152A mutant Filamin A. Effects of Filamin A overexpression: (F) western blot of total protein extracts of IRE1α KO and IRE1α-HA-reconstituted MEFs transfected with plasmid coding for Filamin A-GFP. (G) Left panel: Western blot of total protein extracts of MEFs pretreated with kinases inhibitors IPA3 (5 µM), Gö6976 (100 nM), Ryuvidine (20 µM) and U0126 (5 µM) for 1 h and then treated 100 ng/mL of Tm for 2 h (n = 3 independent experiments). (H) Western blot of total protein extracts of MEFs transfected with 30 pmol of siRNAs for PKCα and PAK1. (I) Western blot of total protein extracts of cells as described in Fig. 5i. Effects of IRE1α expression on PKCα activation: (J) IRE1α KO and IRE1α-HA-reconstituted MEFs were treated with 3% FBS or 100 ng/mL of Tm and analyzed by western blot. (K) Western blot of total protein extracts of IRE1α KO cells transfected with 30 pmol of siRNAs for PKCα or PAK1 treated with 250 ng/mL Tm for 2 h. Control experiments for ER fractionation: (L) Fraction described in 5L were blotted for indicated proteins. In all panels, data represents mean ± s.e.m.; one-way ANOVA followed by Tukey’s test was used, otherwise is indicated; *p < 0.05 and **p < 0.01. Blots represent one out of two (F,J), three (A, I, K, L) experiments with similar results.
Supplementary Figure 6 IRE1α controls actin cytoskeleton dynamics and cell migration independent of its enzymatic activities.
(A) MEFs were co-treated with 100 ng ml–1 of Tm in addition to 25 μM of an IRE1α RNAse specific inhibitor (3-methoxy-6-bromosalicylaldehyde) for 16 h. Xbp1 mRNA splicing was then monitored by RT-PCR. Boyden chamber assay and wound healing assay of MEFs treated with 25 µM of the IRE1α RNAse inhibitor using DMSO as control (n=3 independent experiments). (B) The expression levels of IRE1α of cells from Fig. 6b were analyzed by western blot using an anti-IRE1α or anti-HA antibody. HSP90 levels were monitored as loading control. (C) Left panel: western blot of MEF cells were transiently transfected with expression vectors for IRE1α-HA WT, K599A, P830L, D123P IRE1α-HA or empty vector. Right panel: Boyden chamber assay of cells overexpressing IRE1α WT and mutants described. The number of GFP positive cells that migrated into the lower chamber was counted by fluorescence microscopy (n=4 independent experiments). (D) Upper panel: IRE1α KO MEFs expressing IRE1α-HA WT or the D123P point mutant transfected Life-Act were recorded by time-lapse confocal microscopy every 30 s for 5 min. Segmented images are shown. Lower panel: Average signal over time of the protruding and retracting cell area of cells from Figure S6D was quantified. Filopodia number per cell was calculated using ADAPT software (20 (IRE1α WT) and 15 (IRE1αD123P) independent set of videos.). Control experiments to monitor IRE1α clustering: (E) Upper panel: TREX cells expressing IRE1-3F6H-GFP WT or IRE1-3F6H-GFP D123P were treated with 500 ngml–1 of doxycycline and stimulated with 1 µg/mL of Tm 2 h. Images were obtained by confocal microscopy. Lower panel: quantification of the number of IRE1α cluster per cell (n=2 independent experiments). In all panels, data represents mean ± s.e.m.; one-way ANOVA followed by Tukey’s test was used, otherwise is indicated; *p < 0.05 and **p < 0.01.
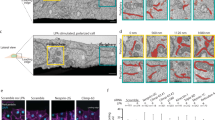
Supplementary Figure 7 IRE1α expression enhances cell migration in fly and zebrafish models in vivo.
(A) Schematic representation of the Mosaic Analysis with a Repressible Cell Marker (MARCM) system. The essence of MARCM is to couple cell division with the loss of a transcriptional repressor (Gal80) in one of the two daughter cells, thus allowing (Gal4/UAS driven) marker expression in one, but not the other, daughter cell, nor in any parental cells. The loss of the transcriptional repressor in one of the daughter cells is achieved by site-specific recombinase FLP/FRT-mediated inter-chromosomal recombination prior to cell division. This system generates IRE1α insertional mutant cells with GFP expression. Control experiments in zebrafish: (B) To control the ability of IRE1α-DN to block endogenous IRE1α signaling in vivo, control and IRE1α-DN injected embryos were treated 18 hpf with 5 μM of thapsigargin for 2 h. Xbp1 mRNA splicing was then monitored by RT–PCR using mRNA purified from total embryonic extracts (n=9). (C) To control the ability of IRE1α-DN to block endogenous IRE1α and Filamin A phosphorylation, HEK cells were transfected with control and IRE1α-DN and treated with 500 ng/mL of Tm for 2 h. Total proteins extracts were analyzed by SDS-PAGE and western blot using specific antibodies for pS2152-Filamin A, Filamin A and HSP90 (n=2 independent experiments). Control experiments for overexpression in zebrafish: (D) global phenotype of embryos injected with IRE1α WT at 40 hpf. Raw data of fig. 8J: (E) plots of average intensity signal taken in lines of 4 µm-length along the cell cortex of individual cells (N = 26 control; N = 25 IRE1α-DN). In all panels, the data are shown as mean ± s.e.m.; one-way ANOVA followed by Tukey’s test was used, otherwise is indicated; *p < 0.05 and **p < 0.01.
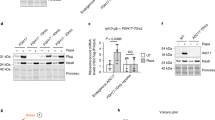
Supplementary Figure 8 IRE1α regulates neuronal cell migration in vitro and in vivo.
IRE1α regulates cell migration in neuronal cells: (A) Left panel: Western blot of total protein extracts and Xbp1 mRNA splicing of IRE1α knockout cells were generated using CRISPR/Cas9in the neuronal cell line HT22.Right panel: Boyden chamber assay of HT22 control or CRISPR was assessed after 6 h has described previously (n=8 independent experiments). Schematic representation of the in utero electroporation protocol: (B) In utero electroporation was performed at E14.5 to deliver shRNA Luc and control or an shRNA against IRE1α mRNA. For comparison, an shRNA targeting Filamin A mRNA was used (Modified from dal Maschio et al43). Expression of IRE1α and Filamin A during brain development: (C) Filamin A or Ire1α mRNAs were quantified by real-time PCR in dissected brain samples obtained from different embryonic stages (E12.5, E14.5 and E17.5), in addition to birth (P0), young age (P15), and adult age mice (AD, > 2 months). mRNA levels were normalized with actin expression. (D) Total proteins extract of samples described in C were analyzed by Western blot using specific antibodies of Filamin A, IRE1α and actin as loading control (n=2 independent experiments). IRE1α expression regulates neuronal migration during brain development upstream of Filamin A: (F) Quantification of the percentage of GFP-positive cells after in utero electroporation in the ventricular zone/subventricular zone (VZ/SVZ), intermediate zone (IZ), layer VI (VI), layers IV/V (IV/V) and layers II/III (II/III) are shown of brains electroporated using constructs for shRNA IRE1α and Filamin A WT-MYC, Filamin A S2152A-MYC or empty vector. These experiments are part of Fig. 7d,e. (G) Individual dot plots of the analysis by cortical layer are shown for all animal groups of experiments presented in 7D. Each circle represents an individual animal (Mock n = 14, Filamin A WT-MYC n = 11 and Filamin A S2152A-MYC n = 7). In all panels, data represents mean ± s.e.m.; one-way ANOVA followed by Tukey’s test was used, otherwise is indicated; *p < 0.05 and **p < 0.01.
Supplementary Figure 9
Scans of all unprocessed blots
Supplementary information
Supplementary Information
Supplementary Figures 1–9, Supplementary Table and Supplementary Video Legends
Supplementary Video 1
IRE1α regulates actin cytoskeleton dynamics
Supplementary Video 2
IRE1α controls actin hemocytes migration in flies
Supplementary Video 3
Interfering IRE1α expression reduces actin cytoskeleton dynamics in D. melanogaster
Supplementary Video 4
IRE1α contributes to cell migration during zebrafish development
Supplementary Video 5
Targeting IRE1α impairs actin cytoskeleton dynamics during zebra fish development
Rights and permissions
About this article
Cite this article
Urra, H., Henriquez, D.R., Cánovas, J. et al. IRE1α governs cytoskeleton remodelling and cell migration through a direct interaction with filamin A. Nat Cell Biol 20, 942–953 (2018). https://doi.org/10.1038/s41556-018-0141-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0141-0
This article is cited by
-
Intracellular remodeling associated with endoplasmic reticulum stress modifies biomechanical compliance of bladder cells
Cell Communication and Signaling (2023)
-
FRMD3 inhibits the growth and metastasis of breast cancer through the ubiquitination-mediated degradation of vimentin and subsequent impairment of focal adhesion
Cell Death & Disease (2023)
-
Distinct Epileptogenic Mechanisms Associated with Seizures in Wolf-Hirschhorn Syndrome
Molecular Neurobiology (2022)
-
Carrying Excess Baggage Can Slowdown Life: Protein Clearance Machineries That Go Awry During Aging and the Relevance of Maintaining Them
Molecular Neurobiology (2022)
-
Endoplasmic reticulum stress signals in the tumour and its microenvironment
Nature Reviews Cancer (2021)