Abstract
The essential ATP-binding cassette protein ABCE1 splits 80S ribosomes into 60S and 40S subunits after canonical termination or quality-control-based mRNA surveillance processes. However, the underlying splitting mechanism remains enigmatic. Here, we present a cryo-EM structure of the yeast 40S–ABCE1 post-splitting complex at 3.9-Å resolution. Compared to the pre-splitting state, we observe repositioning of ABCE1's iron-sulfur cluster domain, which rotates 150° into a binding pocket on the 40S subunit. This repositioning explains a newly observed anti-association activity of ABCE1. Notably, the movement implies a collision with A-site factors, thus explaining the splitting mechanism. Disruption of key interactions in the post-splitting complex impairs cellular homeostasis. Additionally, the structure of a native post-splitting complex reveals ABCE1 to be part of the 43S initiation complex, suggesting a coordination of termination, recycling, and initiation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others

References
Schmitt, L. & Tampé, R. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 12, 754–760 (2002).
Dean, M. & Annilo, T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet. 6, 123–142 (2005).
Rees, D.C., Johnson, E. & Lewinson, O. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10, 218–227 (2009).
Hopfner, K.P. Invited review: architectures and mechanisms of ATP binding cassette proteins. Biopolymers 105, 492–504 (2016).
Jackson, R.J., Hellen, C.U. & Pestova, T.V. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 86, 45–93 (2012).
Franckenberg, S., Becker, T. & Beckmann, R. Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Curr. Opin. Struct. Biol. 22, 786–796 (2012).
Nürenberg, E. & Tampé, R. Tying up loose ends: ribosome recycling in eukaryotes and archaea. Trends Biochem. Sci. 38, 64–74 (2013).
Shoemaker, C.J. & Green, R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 19, 594–601 (2012).
Barthelme, D. et al. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J. Biol. Chem. 282, 14598–14607 (2007).
Karcher, A., Schele, A. & Hopfner, K.P. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi . J. Biol. Chem. 283, 7962–7971 (2008).
Becker, T. et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506 (2012).
Preis, A. et al. Cryoelectron microscopic structures of eukaryotic translation termination complexes containing eRF1-eRF3 or eRF1-ABCE1. Cell Rep. 8, 59–65 (2014).
Brown, A., Shao, S., Murray, J., Hegde, R.S. & Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496 (2015).
Pisarev, A.V. et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 37, 196–210 (2010).
Shoemaker, C.J. & Green, R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. USA 108, E1392–E1398 (2011).
Pisareva, V.P., Skabkin, M.A., Hellen, C.U., Pestova, T.V. & Pisarev, A.V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30, 1804–1817 (2011).
van den Elzen, A.M., Schuller, A., Green, R. & Séraphin, B. Dom34-Hbs1 mediated dissociation of inactive 80S ribosomes promotes restart of translation after stress. EMBO J. 33, 265–276 (2014).
Barthelme, D. et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc. Natl. Acad. Sci. USA 108, 3228–3233 (2011).
Guydosh, N.R. & Green, R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell 156, 950–962 (2014).
Young, D.J., Guydosh, N.R., Zhang, F., Hinnebusch, A.G. & Green, R. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo . Cell 162, 872–884 (2015).
Dong, J. et al. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 279, 42157–42168 (2004).
Andersen, D.S. & Leevers, S.J. The essential Drosophila ATP-binding cassette domain protein, pixie, binds the 40 S ribosome in an ATP-dependent manner and is required for translation initiation. J. Biol. Chem. 282, 14752–14760 (2007).
Skabkin, M.A., Skabkina, O.V., Hellen, C.U. & Pestova, T.V. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell 51, 249–264 (2013).
Hinnebusch, A.G. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31, 553–562 (2006).
Fernández, I.S. et al. Molecular architecture of a eukaryotic translational initiation complex. Science 342, 1240585 (2013).
Behrmann, E. et al. Structural snapshots of actively translating human ribosomes. Cell 161, 845–857 (2015).
Taylor, D.J. et al. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 26, 2421–2431 (2007).
Spahn, C.M. et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 23, 1008–1019 (2004).
Becker, T. et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 18, 715–720 (2011).
Kiosze-Becker, K. et al. Structure of the ribosome post-recycling complex probed by chemical cross-linking and mass spectrometry. Nat. Commun. 7, 13248 (2016).
Oldham, M.L. & Chen, J. Snapshots of the maltose transporter during ATP hydrolysis. Proc. Natl. Acad. Sci. USA 108, 15152–15156 (2011).
Korkhov, V.M., Mireku, S.A., Veprintsev, D.B. & Locher, K.P. Structure of AMP-PNP-bound BtuCD and mechanism of ATP-powered vitamin B12 transport by BtuCD-F. Nat. Struct. Mol. Biol. 21, 1097–1099 (2014).
Villa, E. et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl. Acad. Sci. USA 106, 1063–1068 (2009).
Groft, C.M., Beckmann, R., Sali, A. & Burley, S.K. Crystal structures of ribosome anti-association factor IF6. Nat. Struct. Biol. 7, 1156–1164 (2000).
Tsuboi, T. et al. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell 46, 518–529 (2012).
Lill, R. Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 (2009).
Coelho, C.M. et al. Growth and cell survival are unevenly impaired in pixie mutant wing discs. Development 132, 5411–5424 (2005).
Sauer, R.T. et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119, 9–18 (2004).
Monroe, N. & Hill, C.P. Meiotic clade AAA ATPases: protein polymer disassembly machines. J. Mol. Biol. 428, 1897–1911 (2016).
Llácer, J.L. et al. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell 59, 399–412 (2015).
Merrick, W.C. & Hensold, J.O. Analysis of eukaryotic translation in purified and semipurified systems. Curr. Protoc. Cell. Biol. 8, 11.19.1–11.19.26 (2001).
Zheng, S.Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Kimanius, D., Forsberg, B.O., Scheres, S.H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016).
Kucukelbir, A., Sigworth, F.J. & Tagare, H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Ben-Shem, A. et al. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334, 1524–1529 (2011).
Smith, P.C. et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 (2002).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
Fernández, I.S., Bai, X.C., Murshudov, G., Scheres, S.H. & Ramakrishnan, V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157, 823–831 (2014).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Aylett, C.H.S. et al. Structure of a yeast 40S–Eif1–Eif1A–Eif3–Eif3j initiation complex. Nat. Struct. Mol. Biol. 22, 269–271 (2015).
Acknowledgements
The authors thank K. Kiosze-Becker, E. Nürenberg-Goloub, B. Hetzert, C. Le Gal and C. Thomas for helpful suggestions on the manuscript and C. Ungewickell, S. Lange and S. Lamberth for technical assistance. This work was supported by the German Research Council (grants SFB 902 to R.T., SFB 646 to R.B. and T.B., FOR 1805 to R.B., GRK 1721 to R.B.). R.B. acknowledges support by the Center for Integrated Protein Science Munich (CiPS-M) and the European Research Council (Advanced Grants CRYOTRANSLATION). The Cluster of Excellence–Macromolecular Complexes (EXC 115 to R.T.) supported the work. M.G. and C.S. were supported by Boehringer Ingelheim Fonds PhD fellowships. We thank the Leibniz-Rechenzentrum Munich (LRZ) for providing computational services and support.
Author information
Authors and Affiliations
Contributions
M.G., A.H., T.B., R.B. and R.T. designed the study. M.G. developed the preparation of the post-splitting complex and performed all functional assays. M.G. and P.K. conducted the plasmid shuffling experiment. M.G. and S.T. designed the NTPase assays. M.G. and A.H. prepared the EM samples. A.P. and A.H. prepared the initiation complex. A.H. and O.B. collected and A.H. processed the cryo-EM data. C.S., A.H. and T.B. built and refined the model. C.S., T.B., A.H. and M.G. analyzed and interpreted the structures. M.G., T.B., A.H., S.T., R.B. and R.T. wrote the manuscript. R.T. initiated and R.B. and R.T. conceived the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
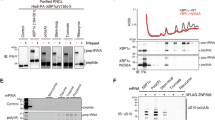
Supplementary Figure 1 Preparation of 40S–ABCE1 post-splitting complex.
a, Outline of the facilitated splitting approach: 80S ribosomes were dissociated under high potassium (500 mM) and low magnesium (1 mM) conditions, followed by AMP‑PNP-dependent ABCE1 binding to the 40S subunit. Under re-association conditions (low potassium and high magnesium concentration (100 mM and 20 mM), free 40S subunits were allowed to rejoin with free 60S subunit. A representative profile of four individual preparations is shown. b, In ribosome profiles, the 40S–ABCE1 complex only appears in the 40S fraction if AMP‑PNP is present and ABCE1 is added under splitting-facilitating conditions. c, Pooled 40S ribosomal fractions show stoichiometric binding of ABCE1 to 40S subunits as analyzed by SDS-PAGE.
Supplementary Figure 2 Raw cryo-EM data and classification of the reconstituted 40S–ABCE1 complex.
a, Representative micrograph showing 40S–ABCE1 particles. b, Representative 2D classes predominantly showing various side views of the 40S subunits. c, 3D classification scheme. After 2D classification and removal of non-ribosomal particles, the dataset was refined and subjected to 3D classification in RELION-2. The dataset was initially classified into four classes: class 1 and 2 contained poorly resolved and distorted 40S ribosomes whereas Class 3 and 4 showed well-resolved 40S ribosomes with a strong ABCE1 density. These classes were joined for a second round of 3D classification (five classes) using a mask excluding the highly flexible 40S head. Four out of five classes showed either a strong distortion which is likely a result of orientation bias or poor alignment and were discarded. The best resolved class 1 showed a strong ABCE1 density and a well resolved 40S body and was used for the refinement yielding a map at 3.9 Å resolution.
Supplementary Figure 3 Cryo-EM structure of the reconstituted 40S–ABCE1 complex and assessment of resolution.
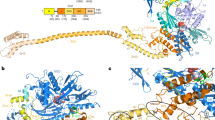
a, Cryo-EM density of the 40S–ABCE1 complex and isolated ABCE1 low-pass filtered at 3.9 Å showing the 40S subunit (grey), and ABCE1 (red), as well as local resolution as calculated by ResMap. The ResMap plots show a range from maximum 3.5 Å to 8.5 Å in the periphery. It is to note, in ABCE1, the FeS cluster domain and NBD1 are well resolved whereas resolution in NBD2 and peripheral regions of ABCE1 is slightly decreased. Maps are contoured at 3.5 σ. b, FSC plot shows the 3.9 Å average resolution of the map according to the “gold standard” criterion (FSC = 0.143; top) and FSC curves calculated between the cryo‑EM map and the final models (bottom) as calculated by REFMAC. Values are plotted for the model versus the final map (FSCaverage, black), for the model that was refined into the first half-map and FSC calculated either for the same map (model vs first half-map, orange) or for the second half-map (model vs second half map, blue). c, Density snapshots of isolated ABCE1 (contoured as indicated in the panels) with the fitted model shown in three orientations. Below, selected areas are shown illustrating the quality of the map (side chain densities in the α-helices forming the HLH motif; a separated β-sheet in NBD1 and a β-sheet with resolved bulky side chains in hinge 2). Domains are colored as indicated in the schematic panel.
Supplementary Figure 4 NTPase stimulation of ABCE1 by 40S subunit.
a, Hydrolyzed γ-32P-GTP resulted in released 32Pi that was separated by thin layer chromatography and quantified by autoradiography. b, Time traces of 32Pi formation were normalized to GTP only and time point 0 min and analyzed by a linear fit. NTP hydrolysis by ABCE1 was stimulated by addition of 40S subunits. c, Corrected GTPase activity of ABCE1 only shows stimulation by 3-fold upon addition of 40S subunit. Data derived from the slope in panel b are given as mean ± s.e.m. (n = 5).
Supplementary Figure 5 Dynamic conformation of the cantilever hinge.
In the pre-splitting state (left, 3JAH)13, the cantilever hinge (CL) forms an α-helix, which is unwound in the ABCE1 post-splitting state (right). Isolated densities are low-pass filtered at 3.9 Å (contoured at 5 σ) and shown with the respective models. b, In the post-splitting state, the FeS cluster domain is mainly stabilized by two interactions of the cantilever arm: the backbone of Asn78 interacts with Tyr301 of NBD1. Density of the ABCE1 post-splitting complex was low-pass filtered at 3.9 Å (contoured at 5 σ).
13 Brown, A., Shao, S., Murray, J., Hegde, R.S. & Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 524, 493-496 (2015).
Supplementary Figure 6 Ribosome profiles from cells expressing wild-type and mutant ABCE1.
a, Lysates from cells expressing wild-type and mutant ABCE1 were subjected to 10-50% sucrose density gradient centrifugation and A260 profiles were recorded. Compared to wild-type (wt, black), the relative amount of 80S ribosome was strongly increased in the alanine mutants R7A, P30A and Y301A. b, SDS-PAGE and immunoblotting of the gradient fractions using an anti-HA antibody to probe for tagged ABCE1. Wild-type ABCE1 was mainly found in the top fractions or bound to 40S subunits, while the mutants R7A, P30A, and Y301A were enriched on 80S ribosomes.
Supplementary Figure 7 Preparation and cryo-EM of the native 40S–ABCE1 complex.
a, The TEV-eluate of affinity purified tandem-affinity tagged ribosome–ABCE1 complexes was applied to a 5-30% sucrose density gradient and the A260 profile was recorded. b, Fractions containing 40S subunits and ABCE1 were pooled and analyzed by SDS-PAGE. Proteins were identified by mass spectrometry. Notably, in addition to ABCE1 and 40S proteins (RPS), the complex also contains components of the 43S pre-initiation complex (eIF2α and eIF2γ, eIF3c and eIF3j) together with the fatty acid synthase as a common contaminant of 40S preparations. c, Samples were subjected to cryo-EM and single-particle analysis. The fatty acid synthase could be easily sorted out during 2D classification. d, After refinement, 3D classification was performed in RELION-2. In the first round three classes (63%) show a clear density for ABCE1 and two of them showed additional extra density emerging from the P-site. These classes (43.5%) were joined for a second round of classification. Here, four of five classes only differ in the appearance of the density in the P-site, which most likely represents initiator tRNA (tRNAi) in various positions. One class, however showed additional density in the position where eIF1A is located. This class (17.6%; 9,500 particles) was refined to a final resolution of 14 Å (e) according to the “gold standard” criterion (FSC = 0.143). The model of ABCE1 in the post-splitting state was fitted as rigid body without further adjustments into the ABCE1 density. eIF1A (blue) as well as tRNAi (green) could be identified by rigid body fitting of 43 and 48S-initiation complex structures (eIF1A from 4UER, ref. 55, tRNAi taken from 3JAP, ref. 40). f, A difference map was calculated between the native (left) and the in vitro reconstituted 40S–ABCE1 maps (middle). The difference map was superimposed to the in vitro reconstituted 40S–ABCE1 map (contoured at 3.5 σ). Notably, significant difference between the two maps (left and middle) occurred in the region of initiator tRNA (tRNAi) and eIF1A. No conformational differences were observed for ABCE1.
40 Llacer, J.L. et al. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell 59, 399-412 (2015).
55 Aylett, C.H. et al. Structure of a yeast 40S–eIF1–eIF1A–eIF3–eIF3j initiation complex. Nat. Struct. Mol. Biol. 22, 269-271 (2015).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1330 kb)
Transition of ABCE1 from the open to the closed state.
In the free state, ABCE1 is in the open state with two ADPs occluded. After binding to 80S pre-termination complexes (with eRF1 or Dom34 or Pelota), ABCE1 changes its conformation to an intermediate, semi-closed (pre-splitting) state. Here the nucleotide occupation is less clear. High and medium resolved cryo-EM structures showed density for a bound nucleotide in NBS I, whereas NBS II was rather empty. Occlusion of both NBSs with ATP leads to closure of the NBDs which is required for ribosome splitting. This closure is accompanied by rearrangement of the FeS cluster domain. In the post-splitting state, ABCE1 can be trapped on the 40S subunit in a closed conformation occluding two ATP (here AMP-PNP). The FeS cluster domain rotates into a binding cleft formed by h5, h44 and uS12. This conformation is mainly stabilized by interactions of Pro30 to uS12 and Arg7 to h5. Moreover, the cantilever helix unwinds and Tyr301 contacts the backbone of Asn78. The HLH motif interacts with the junction of h8 and h14 mainly via Ser150. Additional conformational changes occur in the C-terminal helix of eS24, which contacts NBD1 (Gln262) in post-splitting state. Only minor conformational changes occur for the hinge 2 motif, which contacts the h5 and h15 junction via Arg573 and Ser588. A zoom into NBS I shows how the nucleotide is occluded by ABC motifs. Open (homology model based on PDB: 3BK7), semi-closed (PDB: 3J16), and closed state (this study) of ABCE1 were morphed using UCSF-Chimera. (MP4 117001 kb)
Mechanism of ABCE1-dependent ribosome splitting.
In the pre-splitting state, ABCE1 is bound to an 80S ribosome occupied with an A-site factor (here eRF1). The FeS cluster domain interacts with the C-terminal domain of eRF1 (or Pelota). In phase 1, ATP-binding leads to closing of the NBDs and rearrangement of the FeS cluster domain in direction of the bound A-site factor. It acts as a wedge, which leads to destabilization of the 80S ribosome. In phase 2, after dissociation of the 60S, the FeS cluster domain locks into its final position and prevents formation of an intersubunit bridge B5 involving uL14 (green), thus acting as an anti-association factor. Isolated densities of the 60S subunit (dark grey), eRF1 (blue), pre-splitting state of ABCE1 (yellow) as well as respective models were taken from EMD-2598 (PDB: 4CRM). (MP4 46475 kb)
Rights and permissions
About this article
Cite this article
Heuer, A., Gerovac, M., Schmidt, C. et al. Structure of the 40S–ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat Struct Mol Biol 24, 453–460 (2017). https://doi.org/10.1038/nsmb.3396
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3396
This article is cited by
-
Mechanisms and consequences of mRNA destabilization during viral infections
Virology Journal (2024)
-
Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2
Nature Communications (2023)
-
Pervasive translation of circular RNAs driven by short IRES-like elements
Nature Communications (2022)
-
eIF6 rebinding dynamically couples ribosome maturation and translation
Nature Communications (2022)
-
Iron–sulfur clusters as inhibitors and catalysts of viral replication
Nature Chemistry (2022)