Abstract
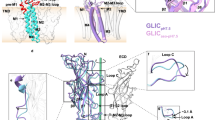
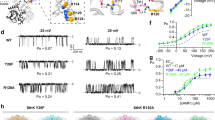
Pentameric ligand-gated ion channels mediate signal transduction through conformational transitions between closed-pore and open-pore states. To stabilize a closed conformation of GLIC, a bacterial proton-gated homolog from Gloeobacter violaceus whose open structure is known, we separately generated either four cross-links or two single mutations. We found all six mutants to be in the same 'locally closed' conformation using X-ray crystallography, sharing most of the features of the open form but showing a locally closed pore as a result of a concerted bending of all of its M2 helices. The mutants adopt several variant conformations of the M2-M3 loop, and in all cases an interacting lipid that is observed in the open form disappears. A single cross-linked mutant is functional, according to electrophysiology, and the locally closed structure of this mutant indicates that it has an increased flexibility. Further cross-linking, accessibility and molecular dynamics data suggest that the locally closed form is a functionally relevant conformation that occurs during allosteric gating transitions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Changeux, J.-P. & Edelstein, S.J. Allosteric mechanisms of signal transduction. Science 308, 1424–1428 (2005).
Burzomato, V., Beato, M., Groot-Kormelink, P.J., Colquhoun, D. & Sivilotti, L.G. Single channel behaviour of heteromeric α1β glycine receptors: an attempt to detect conformational change before the channel opens. J. Neurosci. 24, 10924–10940 (2004).
Mukhtasimova, N., Lee, W.Y., Wang, H.L. & Sine, S.M. Detection and trapping of intermediate states priming nicotinic receptor channel opening. Nature 459, 451–454 (2009).
Hilf, R.J.C. & Dutzler, R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 (2008).
Bocquet, N. et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 (2009).
Hilf, R.J.C. & Dutzler, R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118 (2009).
Hibbs, R.E. & Gouaux, E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature 474, 54–60 (2011).
Brejc, K. et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276 (2001).
Miyazawa, A., Fujiyoshi, Y. & Unwin, N. Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955 (2003).
Giraudat, J., Dennis, M., Heidmann, T., Chang, J.Y. & Changeux, J.P. Structure of the high-affinity binding site for noncompetitive blockers of the acetylcholine receptor: serine-262 of the δ subunit is labeled by [3H]chlorpromazine. Proc. Natl. Acad. Sci. USA 83, 2719–2723 (1986).
Cymes, G.D., Ying, N. & Grosman, C. Probing ion-channel pores one proton at a time. Nature 438, 975–980 (2005).
Bocquet, N. et al. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445, 116–119 (2007).
Nury, H. et al. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature 469, 428–431 (2011).
Baenziger, J.E. & Corringer, P.-J. 3Dstructure and allosteric modulation of the transmembrane domain of pentameric ligand-gated ion channels. Neuropharmacology 60, 116–125 (2011).
Brannigan, G., Hénin, J., Law, R., Eckenhoff, R. & Klein, M.L. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA 105, 14418–14423 (2008).
Wang, H.-L., Chang, X. & Sine, S.M. Intra-membrane proton binding site linked to activation of a bacterial pentameric ion channel. J. Biol. Chem. 287, 6482–6489 (2012).
Nury, H. et al. One-microsecond molecular dynamics simulation of channel gating in a nicotinic receptor homologue. Proc. Natl. Acad. Sci. USA 107, 6275–6280 (2010).
Franklin, J., Koehl, P., Doniach, S. & Delarue, M. MinActionPath: maximum likelihood trajectory for large-scale structural transitions in a coarse-grained locally harmonic energy landscape. Nucleic Acids Res. 35, W477–W482 (2007).
Parikh, R.B., Bali, M. & Akabas, M.H. Structure of M2 transmembrane segment of GLIC, a prokaryotic Cys-loop receptor homologue from Gloeobacter violaceus, probed by substituted cysteine accessibility. J. Biol. Chem. 286, 14098–14109 (2011).
Revah, F. et al. Mutations in the channel domain alter desensitization of a neuronal nicotinic receptor. Nature 353, 846–849 (1991).
Chang, Y. & Weiss, D.S. Substitutions of the highly conserved M2 leucine create spontaneously opening ρ1 γ-aminobutyric acid receptors. Mol. Pharmacol. 53, 511–523 (1998).
Lobo, I.A., Trudell, J.R. & Harris, R.A. Cross-linking of glycine receptor transmembrane segments two and three alters coupling of ligand binding with channel opening. J. Neurochem. 90, 962–969 (2004).
Williams, D.B. & Akabas, M.H. γ-aminobutyric acid increases the water accessibility of M3 membrane-spanning segment residues in γ-aminobutyric acid type A receptors. Biophys. J. 77, 2563–2574 (1999).
Duret, G. et al. Functional prokaryotic-eukaryotic chimera from the pentameric ligand-gated ion channel family. Proc. Natl. Acad. Sci. USA 108, 12143–12148 (2011).
Yamodo, I.H., Chiara, D.C., Cohen, J.B. & Miller, K.W. Conformational changes in the nicotinic acetylcholine receptor during gating and desensitization. Biochemistry 49, 156–165 (2010).
Colquhoun, D. & Sakmann, B. Fast events in single-channel currents activated by acetylcholine and its analogues at the frog muscle end-plate. J. Physiol. (Lond.) 369, 501–557 (1985).
Purohit, P., Mitra, A. & Auerbach, A. A stepwise mechanism for acetylcholine receptor channel gating. Nature 446, 930–933 (2007).
Li, G.-D. et al. Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J. Neurosci. 26, 11599–11605 (2006).
Mihic, S.J. et al. Sites of alcohol and volatile anaesthetic action on GABA(A) and glycine receptors. Nature 389, 385–389 (1997).
Young, G.T., Walker, A.S., Sher, E. & Millar, N.S. Potentiation of α7 nicotinic acetylcholine receptors via an allosteric transmembrane site. Proc. Natl. Acad. Sci. USA 105, 14686–14691 (2008).
Taly, A., Corringer, P.J., Guedin, D., Lestage, P. & Changeux, J.P. Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nat. Rev. Drug Discov. 8, 733–750 (2009).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of 21 initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Blanc, E., Roversi, P., Vonrhein, C., Flensburg, S.M. & Bricogne, G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Brooks, B.R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009).
Krieger, E., Nielsen, J.E., Spronk, C.A. & Vriend, G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 25, 481–486 (2006).
Phillips, J.C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald - an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Tuckerman, M., Berne, B.J. & Martyna, G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 97, 1990–2001 (1992).
Tama, F. & Sanejouand, Y.-H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 14, 1–6 (2001).
PyMOL. (The PyMOL Molecular Graphics System, Version 1.2r3pre) (Schrödinger, LLC, version 0.99, 2006).
Smart, O.S., Goodfellow, J.M. & Wallace, B.A. The pore dimensions of gramicidin A. Biophys. J. 65, 2455–2460 (1993).
Acknowledgements
This work was supported by the European Commission grant 'NeuroCypres' to M.D. and P.-J.C., and the Louis D. Foundation from the Institut de France (to P.-J.C.). Molecular simulations were performed using high performance computing resources from Grand Equipement National de Calcul Intensif (GENCI)–Institut du Développement et des Ressources en Informatique Scientifique (IDRIS) (grant 2010-072292 to M.D.). The authors thank the staff at Soleil (PXl) and European Synchotron Radiation Facility (ESRF) (especially at beamlines ID29, ID14-4) for their help and their useful advice during X-ray Data Collection. We thank T. Allen for helping us optimize the setup of the simulation system. We thank J.-P. Changeux, S. Edelstein, A. Taly and D. Digregorio for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
M.S.P. carried out mutagenesis, electrophysiology, immunofluorescence and biochemistry with the help of C.V.R. and C.H. L.S. and H.N. carried out protein production, crystal growth, data collection and model refinement. L.S. and H.N. performed structure analysis with the help of F.P. and M.S.P. F.P. carried out the normal mode analysis. M.B. carried out molecular dynamic simulations, which were analyzed by M.B. and F.P. M.D. and P.-J.C. supervised the work. M.S.P. and P.-J.C. wrote the manuscript with the help of the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–3 (PDF 2387 kb)
Supplementary Video 1
MinActionPath plausible trajectory between the Open and LC conformation at the Cα level of description, according to Supplementary Fig. 5. The movie shows the TMD alone in upper view. (MPG 2829 kb)
Rights and permissions
About this article
Cite this article
Prevost, M., Sauguet, L., Nury, H. et al. A locally closed conformation of a bacterial pentameric proton-gated ion channel. Nat Struct Mol Biol 19, 642–649 (2012). https://doi.org/10.1038/nsmb.2307
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2307
This article is cited by
-
A release of local subunit conformational heterogeneity underlies gating in a muscle nicotinic acetylcholine receptor
Nature Communications (2024)
-
Untangling Direct and Domain-Mediated Interactions Between Nicotinic Acetylcholine Receptors in DHA-Rich Membranes
The Journal of Membrane Biology (2019)
-
Prediction and validation of protein intermediate states from structurally rich ensembles and coarse-grained simulations
Nature Communications (2016)
-
X-ray structure of the mouse serotonin 5-HT3 receptor
Nature (2014)
-
General rules for the arrangements and gating motions of pore-lining helices in homomeric ion channels
Nature Communications (2014)