Abstract
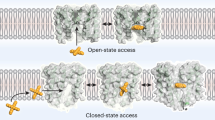
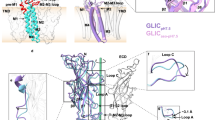
The flow of ions through cation-selective members of the pentameric ligand-gated ion channel family is inhibited by a structurally diverse class of molecules that bind to the transmembrane pore in the open state of the protein. To obtain insight into the mechanism of channel block, we have investigated the binding of positively charged inhibitors to the open channel of the bacterial homolog GLIC by using X-ray crystallography and electrophysiology. Our studies reveal the location of two regions for interactions, with larger blockers binding in the center of the membrane and divalent transition metal ions binding to the narrow intracellular pore entry. The results provide a structural foundation for understanding the interactions of the channel with inhibitors that is relevant for the entire family.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hille, B. Ion Channels of Excitable Membranes 3rd edn. (Sinauer Associates Inc., Sunderland, Massachusetts, USA, 2001).
Sine, S.M. & Engel, A.G. Recent advances in Cys-loop receptor structure and function. Nature 440, 448–455 (2006).
Corringer, P.J., Le Novere, N. & Changeux, J.P. Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 40, 431–458 (2000).
Karlin, A. Emerging structure of the nicotinic acetylcholine receptors. Nat. Rev. Neurosci. 3, 102–114 (2002).
Adler, M., Oliveira, A.C., Albuquerque, E.X., Mansour, N.A. & Eldefrawi, A.T. Reaction of tetraethylammonium with the open and closed conformations of the acetylcholine receptor ionic channel complex. J. Gen. Physiol. 74, 129–152 (1979).
Leonard, R.J., Labarca, C.G., Charnet, P., Davidson, N. & Lester, H.A. Evidence that the M2 membrane-spanning region lines the ion channel pore of the nicotinic receptor. Science 242, 1578–1581 (1988).
Revah, F. et al. The noncompetitive blocker [3H]chlorpromazine labels three amino acids of the acetylcholine receptor gamma subunit: implications for the alpha-helical organization of regions MII and for the structure of the ion channel. Proc. Natl. Acad. Sci. USA 87, 4675–4679 (1990).
Charnet, P. et al. An open-channel blocker interacts with adjacent turns of alpha-helices in the nicotinic acetylcholine receptor. Neuron 4, 87–95 (1990).
Pascual, J.M. & Karlin, A. Delimiting the binding site for quaternary ammonium lidocaine derivatives in the acetylcholine receptor channel. J. Gen. Physiol. 112, 611–621 (1998).
Neher, E. & Steinbach, J.H. Local anaesthetics transiently block currents through single acetylcholine-receptor channels. J. Physiol. (Lond.) 277, 153–176 (1978).
Nutter, T.J. & Adams, D.J. Monovalent and divalent cation permeability and block of neuronal nicotinic receptor channels in rat parasympathetic ganglia. J. Gen. Physiol. 105, 701–723 (1995).
Adams, D.J., Dwyer, T.M. & Hille, B. The permeability of endplate channels to monovalent and divalent metal cations. J. Gen. Physiol. 75, 493–510 (1980).
Dani, J.A. & Eisenman, G. Monovalent and divalent cation permeation in acetylcholine receptor channels. Ion transport related to structure. J. Gen. Physiol. 89, 959–983 (1987).
Hilf, R.J. & Dutzler, R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379 (2008).
Hilf, R.J. & Dutzler, R. A prokaryotic perspective on pentameric ligand-gated ion channel structure. Curr. Opin. Struct. Biol. 19, 418–424 (2009).
Hilf, R.J. & Dutzler, R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457, 115–118 (2009).
Bocquet, N. et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457, 111–114 (2009).
Bocquet, N. et al. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature 445, 116–119 (2007).
Imoto, K. et al. Rings of negatively charged amino acids determine the acetylcholine receptor channel conductance. Nature 335, 645–648 (1988).
Konno, T. et al. Rings of anionic amino acids as structural determinants of ion selectivity in the acetylcholine receptor channel. Proc. Biol. Sci. 244, 69–79 (1991).
Woodhull, A.M. Ionic blockage of sodium channels in nerve. J. Gen. Physiol. 61, 687–708 (1973).
Gumilar, F., Arias, H.R., Spitzmaul, G. & Bouzat, C. Molecular mechanisms of inhibition of nicotinic acetylcholine receptors by tricyclic antidepressants. Neuropharmacology 45, 964–976 (2003).
Sepúlveda, M.I., Baker, J. & Lummis, S.C. Chlorpromazine and QX222 block 5–HT3 receptors in N1E–115 neuroblastoma cells. Neuropharmacology 33, 493–499 (1994).
Yu, Y., Shi, L. & Karlin, A. Structural effects of quinacrine binding in the open channel of the acetylcholine receptor. Proc. Natl. Acad. Sci. USA 100, 3907–3912 (2003).
Armstrong, C.M. Interaction of tetraethylammonium ion derivatives with the potassium channels of giant axons. J. Gen. Physiol. 58, 413–437 (1971).
Hille, B. Common mode of action of three agents that decrease the transient change in sodium permeability in nerves. Nature 210, 1220–1222 (1966).
Weisstaub, N., Vetter, D.E., Elgoyhen, A.B. & Katz, E. The alpha9alpha10 nicotinic acetylcholine receptor is permeable to and is modulated by divalent cations. Hear. Res. 167, 122–135 (2002).
Bertrand, D., Galzi, J.L., Devillers-Thiéry, A., Bertrand, S. & Changeux, J.P. Mutations at two distinct sites within the channel domain M2 alter calcium permeability of neuronal alpha 7 nicotinic receptor. Proc. Natl. Acad. Sci. USA 90, 6971–6975 (1993).
Imoto, K. et al. Location of a delta-subunit region determining ion transport through the acetylcholine receptor channel. Nature 324, 670–674 (1986).
Wilson, G.G., Pascual, J.M., Brooijmans, N., Murray, D. & Karlin, A. The intrinsic electrostatic potential and the intermediate ring of charge in the acetylcholine receptor channel. J. Gen. Physiol. 115, 93–106 (2000).
Lenaeus, M.J., Vamvouka, M., Focia, P.J. & Gross, A. Structural basis of TEA blockade in a model potassium channel. Nat. Struct. Mol. Biol. 12, 454–459 (2005).
Heravi, M.M., Abdolhosseini, N. & Oskooie, H.A. Regioselective and high-yielding bromination of aromatic compounds using hexamethylenetetramine–bromine. Tetrahedr. Lett. 46, 8959–8963 (2005).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Collaborative Computational Project Number 4. The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Kleywegt, G. & Jones, T.A. In Proceedings of the CCP4 Study Weekend. (eds. Bailey, S., Hubbard, R. & Waller, D.), 59–66 (Daresbury Laboratory, Daresbury, UK, 1994).
Sanner, M.F., Olson, A.J. & Spehner, J.C. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305–320 (1996).
Lorenz, C., Pusch, M. & Jentsch, T.J. Heteromultimeric CLC chloride channels with novel properties. Proc. Natl. Acad. Sci. USA 93, 13362–13366 (1996).
Brooks, B.R. et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983).
Im, W., Beglov, D. & Roux, B. Continuum solvation model: computation of electrostatic forces from numerical solutions to the Poisson-Bolztmann equation. Comput. Phys. Commun. 111, 59–75 (1998).
Acknowledgements
We would like to thank the staff of the X06SA beamline for support during data collection and members of the Dutzler lab for help in all stages of the project. Data collection was done at the Swiss Light Source of the Paul Scherrer Institute. The research leading to these results received funding from a grant from the Swiss National Science Foundation (SNF) and from an EC FP7 grant for the European Drug Initiative on Channels and Transporters consortium (HEALTH-201924). R.J.C.H. received the support of the Forschungskredit of the University of Zurich. C.B. and I.Z. are affiliated with the Biomolecular Structure and Mechanism PhD program of the University of Zurich (UZH) and the Swiss Federal Institute of Technology (ETH) Zurich.
Author information
Authors and Affiliations
Contributions
R.J.C.H. and C.B. carried out all experiments. I.Z. assisted in electrophysiological and crystallographic data collection. A.R. and D.T. synthesized the channel blockers. R.D., R.J.C.H. and C.B. jointly planned the experiments and analyzed the data. R.D. wrote the manuscript with the help of all coauthors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Table 1 (PDF 702 kb)
Rights and permissions
About this article
Cite this article
Hilf, R., Bertozzi, C., Zimmermann, I. et al. Structural basis of open channel block in a prokaryotic pentameric ligand-gated ion channel. Nat Struct Mol Biol 17, 1330–1336 (2010). https://doi.org/10.1038/nsmb.1933
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1933
This article is cited by
-
Mechanisms of inhibition and activation of extrasynaptic αβ GABAA receptors
Nature (2022)
-
Prediction and validation of protein intermediate states from structurally rich ensembles and coarse-grained simulations
Nature Communications (2016)
-
Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC
Scientific Reports (2015)
-
Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels
The EMBO Journal (2013)
-
Mass spectrometry of intact membrane protein complexes
Nature Protocols (2013)