Abstract
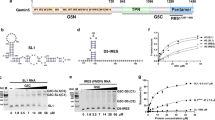
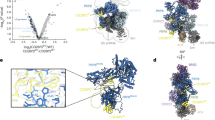
Gemin5 binds specifically to the small nuclear RNA (snRNA)-defining small nuclear ribonucleoprotein (snRNP) code sequence and is essential, together with other components of the survival of motor neurons (SMN) complex, for the biogenesis of snRNPs, the major constituents of spliceosomes. We show that this binding is mediated by Gemin5's WD repeat domain, a common domain not previously known to bind RNA independently. The entire WD repeat domain, comprising 13 WD motifs, is both necessary and sufficient for sequence-specific, high-affinity binding of Gemin5 to its RNA targets. Using an RNA-mediated hydroxyl radical probing method and mass spectrometry, we mapped a discrete region of the WD repeat domain that contacts snRNAs and demonstrated by mutagenesis that specific amino acids in this region are crucial for Gemin5-snRNA binding. The WD repeat domain is thus a previously undescribed RNA binding domain, and we suggest that the presence of WD repeats should be considered as predictive of potential function in RNA binding.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Pellizzoni, L., Yong, J. & Dreyfuss, G. Essential role for the SMN complex in the specificity of snRNP assembly. Science 298, 1775–1779 (2002).
Fischer, U., Liu, Q. & Dreyfuss, G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90, 1023–1029 (1997).
Meister, G., Buhler, D., Pillai, R., Lottspeich, F. & Fischer, U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3, 945–949 (2001).
Will, C.L. & Luhrmann, R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290–301 (2001).
Carissimi, C. et al. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J. Biol. Chem. 281, 8126–8134 (2006).
Liu, Q., Fischer, U., Wang, F. & Dreyfuss, G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90, 1013–1021 (1997).
Charroux, B. et al. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 147, 1181–1194 (1999).
Charroux, B. et al. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 148, 1177–1186 (2000).
Baccon, J., Pellizzoni, L., Rappsilber, J., Mann, M. & Dreyfuss, G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 277, 31957–31962 (2002).
Gubitz, A.K. et al. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 277, 5631–5636 (2002).
Pellizzoni, L., Baccon, J., Rappsilber, J., Mann, M. & Dreyfuss, G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 277, 7540–7545 (2002).
Carissimi, C. et al. Unrip is a component of SMN complexes active in snRNP assembly. FEBS Lett. 579, 2348–2354 (2005).
Lefebvre, S. et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165 (1995).
Lefebvre, S. et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 16, 265–269 (1997).
Wan, L. et al. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 25, 5543–5551 (2005).
Zhang, Z. et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133, 585–600 (2008).
Raker, V.A., Plessel, G. & Luhrmann, R. The snRNP core assembly pathway: identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15, 2256–2269 (1996).
Yong, J., Wan, L. & Dreyfuss, G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 14, 226–232 (2004).
Battle, D.J. et al. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell 23, 273–279 (2006).
Golembe, T.J., Yong, J. & Dreyfuss, G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol. Cell. Biol. 25, 10989–11004 (2005).
Patel, A.A. & Steitz, J.A. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 4, 960–970 (2003).
Yong, J., Golembe, T.J., Battle, D.J., Pellizzoni, L. & Dreyfuss, G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell. Biol. 24, 2747–2756 (2004).
Lunde, B.M., Moore, C. & Varani, G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 (2007).
Burd, C.G. & Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 (1994).
Neer, E.J., Schmidt, C.J., Nambudripad, R. & Smith, T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300 (1994).
Li, D. & Roberts, R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58, 2085–2097 (2001).
Smith, T.F., Gaitatzes, C., Saxena, K. & Neer, E.J. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185 (1999).
Kent, O.A. & MacMillan, A.M. Early organization of pre-mRNA during spliceosome assembly. Nat. Struct. Biol. 9, 576–581 (2002).
Datwyler, S.A. & Meares, C.F. Protein-protein interactions mapped by artificial proteases: where sigma factors bind to RNA polymerase. Trends Biochem. Sci. 25, 408–414 (2000).
Mohri, K., Vorobiev, S., Fedorov, A.A., Almo, S.C. & Ono, S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279, 31697–31707 (2004).
Kelley, L.A., MacCallum, R.M. & Sternberg, M.J. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299, 499–520 (2000).
Labarga, A., Valentin, F., Andersson, M. & Lopez, R. Web services at the European Bioinfomatics Institute. Nucleic Acids Res. 35, W6–W11 (2007).
Kickhoefer, V.A., Stephen, A.G., Harrington, L., Robinson, M.O. & Rome, L.H. Vaults and telomerase share a common subunit, TEP1. J. Biol. Chem. 274, 32712–32717 (1999).
Lukowiak, A.A. et al. Interaction of the U3–55k protein with U3 snoRNA is mediated by the box B/C motif of U3 and the WD repeats of U3–55k. Nucleic Acids Res. 28, 3462–3471 (2000).
Granneman, S. et al. The hU3–55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in Vitro. J. Biol. Chem. 277, 48490–48500 (2002).
Yong, J., Pellizzoni, L. & Dreyfuss, G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 21, 1188–1196 (2002).
Cohen, S.B. & Cech, T.R. Dynamics of thermal motions within a large catalytic RNA investigated by cross-linking with thiol-disulfide interchange. J. Am. Chem. Soc. 119, 6259–6268 (1997).
Newcomb, L.F. & Noller, H.F. Directed hydroxyl radical probing of 16S rRNA in the ribosome: spatial proximity of RNA elements of the 3′ and 5′ domains. RNA 5, 849–855 (1999).
Voegtli, W.C., Madrona, A.Y. & Wilson, D.K. The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization. J. Biol. Chem. 278, 34373–34379 (2003).
Petrey, D. & Honig, B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 374, 492–509 (2003).
Acknowledgements
We thank the members of our laboratory, especially D. Battle, L. Wan, J. Yong, M. Kasim and I. Younis, for stimulating discussions and helpful comments on this manuscript. We thank S. Pesiridis and K. Lorenz for help with mass spectrometry. This work was supported by the Association Française Contre les Myopathies (AFM). G.D. is funded by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
C-k.L. designed and performed the experiments and wrote the manuscript; J.L.B. performed the experiments; G.D. directed the project, designed experiments and wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1 and Supplementary Tables 1 and 2 (PDF 408 kb)
Rights and permissions
About this article
Cite this article
Lau, Ck., Bachorik, J. & Dreyfuss, G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat Struct Mol Biol 16, 486–491 (2009). https://doi.org/10.1038/nsmb.1584
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1584
This article is cited by
-
Gemin5-dependent RNA association with polysomes enables selective translation of ribosomal and histone mRNAs
Cellular and Molecular Life Sciences (2022)
-
Arabidopsis AtPRP17 functions in embryo development by regulating embryonic patterning
Planta (2021)
-
Loss of function mutations in GEMIN5 cause a neurodevelopmental disorder
Nature Communications (2021)
-
A systematic analysis of the RNA-targeting potential of secreted bacterial effector proteins
Scientific Reports (2017)
-
UsnRNP biogenesis: mechanisms and regulation
Chromosoma (2017)