Abstract
ADP-ribosylation of proteins was first described in the early 1960's, and today the function and regulation of poly(ADP-ribosyl)ation (PARylation) is partially understood. By contrast, little is known about intracellular mono(ADP-ribosyl)ation (MARylation) by ADP-ribosyl transferase (ART) enzymes, such as ARTD10. Recent findings indicate that MARylation regulates signalling and transcription by modifying key components in these processes. Emerging evidence also suggests that specific macrodomain-containing proteins, including ARTD8, macroD1, macroD2 and C6orf130, which are distinct from those affecting PARylation, interact with MARylation on target proteins to 'read' and 'erase' this modification. Thus, studying macrodomain-containing proteins is key to understanding the function and regulation of MARylation.
Similar content being viewed by others
Main
The post-translational modification (PTM) ADP-ribosylation has been studied for five decades and, in addition to the bacterial ADP-ribosyltransferases (ARTs)1,2,3, more than 20 eukaryotic enzymes that can transfer ADP-ribose onto substrates have been identified4,5,6. Eukaryotic ARTs can be subdivided into two groups: those that reside on the plasma membrane and have an extracellular function4,5 and those that are located within the cell. The intracellular enzymes were known as the poly(ADP-ribose)polymerase (PARP) family6,7, but recently, a new nomenclature was proposed to reflect the different catalytic properties and structural features of the enzymes within this family5 (Table 1). Importantly, it has become clear that only a subgroup of the intracellular ARTs can form PAR, whereas the other intracellular enzymes contain an amino acid substitution in their catalytic centre that enables them to attach just mono(ADP-ribose) (MAR) moieties to their targets8. The PARP family has thus been renamed the ARTD family and the extracellular enzymes, which were formerly known as the mART family, were renamed the ARTC family5. These family names take into account the transferase activity of the members, rather than the polymerase activity. Additionally, these names reflect the homology of family members to certain toxins. The structural characterization of the catalytic domains of the ARTs diphtheria toxin and clostridial C2 and C3 toxins revealed that these have distinct folds. The fold in diphtheria toxin is related to the intracellular ARTs and the fold in the clostridial toxins is related to the extracellular ARTs, hence the extension D and C, respectively, to the ART family name5. For ARTD enzymes, it is not entirely clear which amino acid serves as the ADP-ribose acceptor. Some studies indicate that Lys residues are the main acceptor9,10,11, whereas others have identified modified Glu and Asp residues12,13,14. Thus, the amino acid specificity of distinct ARTD enzymes is currently an open question.
The polymer-forming enzymes of the ARTD family, in particular ARTD1 (see Table 1 for a summary of alternative names), ARTD5 and ARTD6, have been reasonably well-studied7,15. Poly(ADP-ribosyl)ation (PARylation) has been shown to influence different signalling processes, and distinct protein modules are known to bind this modification7,15,16. One such module is the macrodomain, which was originally identified in the core histone variant macroH2A17. Four years ago, macrodomains were shown to be able to bind PAR that has been synthesized in response to DNA damage20,25,26. More recently, the macrodomain of PAR glycohydrolase (PARG) was identified as a PAR hydrolase21,22,23, demonstrating that distinct macrodomains can both read and process PAR in addition to their ability to interact with ADP-ribose. We use the term 'reader' throughout this article to indicate proteins that recognize, but do not process, substrates that are modified with ADP-ribose. Furthermore, in our terminology 'writers' and 'erasers' refer to the enzymes that add and remove ADP-ribosylation, respectively.
In contrast to PARylation, MARylation by ARTD family members is less well understood, partially due to the fact that the mechanism of this PTM has only recently been characterized8. However, several reports now indicate that MARylation also has a role in different intracellular signalling processes. Moreover, certain macrodomains have now been identified as binding modules for this PTM and, in addition, other macrodomain-containing proteins have been reported to remove MAR from substrate proteins. These novel findings, which are reviewed in this Opinion article, suggest that MARylation is a PTM that can be read and erased by macrodomain-containing proteins. In light of these observations, we propose that MARylation is a reversible PTM that serves to control protein–protein interactions or enzymatic activity, and that macrodomains are key modules that regulate MARylation. Moreover, it seems probable that additional macrodomain-containing proteins that affect MARylation will be identified and that additional readers and erasers of MARylation that possess different structures also exist.
Functions of PARylation
The modification of substrates by PAR is, compared to many other PTMs, rather unique as it results in the addition of long, negatively charged polymers of ADP-ribose. A considerable number of studies were published in recent years that describe the functional consequences of PARylation. Probably the best-known function of PAR is its role in the DNA damage response in which ARTD1 is recruited to DNA and activated by DNA damage, followed by local PAR synthesis24. These PAR chains then serve as a docking site for several proteins involved in DNA damage repair and the associated chromatin remodelling factors, including APLF, ALC1, CHFR and macroH2A1.1 (Refs 18,19,20,25,26,27). PARylation is also involved in several other processes, including transcriptional regulation, apoptosis, telomere maintenance, mitotic spindle assembly and function, as well as WNT signalling (for recent reviews, see Refs 7,15,16).
In some cases, PARylation directly affects the properties of the modified protein. For example, auto-PARylation of ARTD1 results in its release from DNA28, and PARylation of TRF1 (telomeric repeat-binding factor 1) by ARTD5 inhibits the ability of TRF1 to bind to telomeric DNA29. Interestingly, in many other cases, PAR chains serve as scaffolding modules that attract interacting proteins through specific binding domains. For example, the E3-ubiquitin ligase RING finger 146 (RNF146) binds to the PAR chains generated on axin by ARTD5 through its WWE (that is, Trp-Trp-Glu) domain30, and subsequently ubiquitylates axin. Thus, axin PARylation indirectly promotes its proteasomal degradation31,32. As axin is an inhibitory component of the β-catenin destruction complex in the WNT signalling pathway33, the ART activity of ARTD5 stimulates this signal transduction pathway.
Two further modules known to bind to PAR are the PAR-binding zinc-finger (PBZ), which is found in DNA damage response proteins such as CHFR and APLF19, and the PAR-binding motif (PBM). The PBM is a short peptide sequence with clusters of basic and hydrophobic amino acids, and it was the first motif that was identified to bind PAR34,35. PBMs have been found in a considerable number of proteins, many of which are involved in DNA-associated processes such as DNA repair, replication and chromosome organization. Moreover, some proteins linked to mRNA metabolism and protein biosynthesis possess PBMs35. These PAR-binding modules have recently been reviewed and are therefore not further discussed here7,15,16,36.
The fourth PAR-binding module is the macrodomain. Macrodomains are evolutionarily conserved structural modules of 130–190 amino acids that are found in proteins in many different organisms, and they are associated with several distinct activities (Box 1). With respect to PAR, ALC1 is recruited to sites of DNA damage through its macrodomain, and this is dependent on the activation of PAR-forming ARTDs7,15. Moreover, a recent study revealed that the catalytic domain of PARG, the major enzyme responsible for the catabolism of PAR, possesses a macrodomain fold21,22,23,37. These findings demonstrate that macrodomains are intimately involved in PAR metabolism. With regards to the catabolism of PAR, it is worth noting that a second enzyme, ADP-ribosylhydrolase 3 (ARH3), can remove PARylation38. The structure of the catalytic domain of this protein has a distinct fold without obvious homology to macrodomains39. PARG, and most likely also ARH3, seem to be unable to remove the protein proximal ADP-ribose unit, even though some earlier reports suggested it could. Considering that the bonds between two ADP-ribose units and between ADP-ribose and a protein are different, it is not too surprising that different enzymes are required for these cleavage events. Thus, the activity of PARG on a PARylated substrate results in a MARylated product21,40. In contrast to PARylation, less is known about the functional relevance of MARylation, as well as the structures that read it and the enzymes that reverse this modification. The most recent findings, however, strongly indicate that macrodomains also control the functions of this PTM, that is, that macrodomains function as readers and erasers of MARylation.
Emerging functions of MARylation
It has been postulated, on the basis of sequence similarities and structural predictions, that nine ARTD enzymes possess MARylation activity8 (Table 1). These enzymes have recognizable domains and motifs in addition to the catalytic domain, suggesting that they contribute to diverse cellular processes41. Indeed, very recent findings suggest that these enzymes have defined roles.
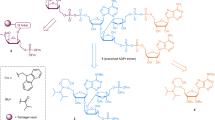
ARTD10 regulates cell signalling. ARTD10 was initially identified as an interaction partner of the transcription factor and proto-oncoprotein MYC42, and it was the first ARTD to be characterized as a mono-ADP-ribosyltransferase8. The nuclear interaction of ARTD10 with MYC was verified, although most ARTD10 seems to be present in highly dynamic cytoplasmic foci that are rich in ubiquitin43. ARTD10 is the only ARTD family member that possesses recognizable ubiquitin-interaction motifs. These bind to Lys63-linked polyubiquitin, which seems to be important for the ability of ARTD10 to repress nuclear factor-κB (NF-κB) signalling44. Within the NF-κB signalling pathway, ARTD10 MARylates NEMO (NF-κB essential modulator), which results in reduced NEMO polyubiquitylation and thus decreased NF-κB signalling (Fig. 1a). It is postulated that the Lys63-linked polyubiquitin chains serve as scaffolds to bring ARTD10 into close proximity to NEMO, which also interacts with the ubiquitin chains45. Moreover, overexpression of ARTD10 leads to apoptosis in HeLa cells dependent on its catalytic activity, and ARTD10 knockdown increases cell survival upon DNA damage caused by doxorubicin46. Because NF-κB signalling promotes cell survival, it is possible that ARTD10 affects apoptosis through repressing this signalling pathway. However, other explanations are also possible.
a | In response to activated receptor complexes the NF-κB essential modulator (NEMO)–IκB kinase (IKK) complex is phosphorylated and ubiquitylated and thereby activated. The ADP-ribosyltransferase ARTD10 mono-(ADP-ribosyl)ates (MARylates) NEMO and reduces its polyubiquitylation, which is dependent on both the ARTD10 ubiquitin-interaction motifs (UIMs) and its catalytic activity. This prevents NEMO from triggering the degradation of inhibitor of κB (IκB), which sequesters NF-κB in the cytoplasm to decrease its nuclear translocation and transcriptional activity. b | ARTD8 functions as an activator of STAT6 (signal transducer and activator of transcription 6), in a manner dependent on its catalytic activity and its macrodomains. Interleukin-4 (IL-4) stimulates the catalytic activity of ARTD8, leading to the MARylation of histone deacetylase 2 (HDAC2) and HDAC3 and their subsequent dissociation from the IL-4-responsive promoter. This allows STAT6 and the transcriptional cofactors p100 and p300 to bind to the promoter. p100 is also MARylated by ARTD8, and this might help bridge STAT6 to the RNA polymerase II (Pol II) complex. One function of p300 is to modify core histones to promote transcription, potentially cooperating with the activity of p100. c | ARTD14 interacts with aryl hydrocarbon receptor (AHR) (which forms a complex with the aryl hydrocarbon receptor nuclear translocator (ARNT)), and represses AHR-mediated transcription dependent on its catalytic domain and zinc-finger domain. Although ARTD14 was shown to MARylate core histones, it is not clear whether histones are the substrates relevant for the repressive effect of ARTD14 on AHR or whether other target proteins play a part. d | ARTD15 is located at the endoplasmic reticulum (ER), where it MARylates the cytosolic part of the ER stress sensors PERK (protein kinase RNA-like ER kinase) and IRE1α (inositol-requiring transmembrane kinase and endonuclease 1α), leading to an increase in their activity. In the ER lumen the carboxyl terminus of ARTD15 is indispensable for PERK and IRE1α activation, potentially because it stimulates the dissociation of the inhibitory factor BiP (binding immunoglobulin protein) from them. ARTD15 thus stimulates the unfolded protein response (UPR). A third ER stress sensor, ATF6, is not modified by ARTD15. TMD, transmembrane domain.
Additional substrates of ARTD10 were identified in protein microarrays, including several kinases47. Glycogen synthase kinase 3β (GSK3β) was investigated to verify the screen and to address the consequences of MARylation. It was found that GSK3β displays reduced kinase activity when it is modified by MAR. Overexpression of ARTD10 decreased GSK3β activity, whereas knockdown of endogenous ARTD10 enhanced GSK3β activity, which suggests that GSK3β MARylation occurs in cells47. MARylation of GSK3β could thus modulate the function of this kinase in downstream signalling pathways, such as WNT signalling, in which GSK3β phosphorylates β-catenin to lead to β-catenin proteasomal degradation48. Together, these findings indicate that ARTD10–mediated MARylation is involved in multiple signalling processes.
ARTD8 and ARTD14 regulate transcription. Substrates of ARTD8 were also identified using protein microarrays, although they were not further characterized47. ARTD8 potentiates STAT6 (signal transducer and activator of transcription 6)-mediated transcription and, in accordance with this, it was named co-activator of STAT6 (CoaSt6) in earlier studies. In these studies, ARTD8 was shown to ADP-ribosylate p100 (Refs 49,50,51) (Fig. 1b), but it was not determined whether p100 was MARylated or PARylated. p100 functions as a STAT6 cofactor, bridging this transcription factor to the RNA polymerase II (Pol II) complex52. A catalytically inactive ARTD8 mutant could not potentiate STAT6 activity, which indicates that catalytic activity is essential. However, whether p100 is the relevant substrate of ARTD8 remains to be determined. Additionally, ARTD8 binds to the transcriptional repressors histone deacetylase 2 (HDAC2) and HDAC3 in unstimulated cells and ADP-ribosylates these deacetylases upon interleukin-4 (IL-4) stimulation. This leads to their dissociation from interleukin-4 (IL-4)-responsive promoters and thus allows binding of transcriptional activators such as STAT6, p300 and possibly p100 (Ref. 50).
Together, these findings suggest that ARTD8 targets multiple proteins to regulate gene transcription, but additional studies are required to fully understand the relationship between individual observations. In a very recent paper, a requirement for ARTD8 in allergic airway disease was described. ARTD8 and its catalytic activity promote T cell differentiation towards a TH2 phenotype53. This work suggests that ARTD8 inhibitors might be of therapeutic relevance for allergic inflammation.
In addition to the ability of ARTD8 to regulate transcription, ARTD14 has been proposed to act as a transcriptional repressor of the aryl hydrocarbon receptor (AHR), in a manner dependent on its zinc-finger and catalytic domain54 (Fig. 1c). Although the catalytic activity of ARTD14 was confirmed to be MARylation of core histones, it is not clear through which substrates it exerts its effect on AHR activity54.
ARTD15 regulates the unfolded protein response. ARTD15 has a unique transmembrane domain that induces its localization to the endoplasmic reticulum (ER) and the nuclear envelope55,56. A mass spectrometry-based search for ARTD15-interacting partners identified karyopherin β1 (KAPβ1), which is a nuclear transport protein. Subsequently, KAPβ1 was found to be also a substrate of ARTD15 (Ref. 56). The functional relevance of this ARTD15–KAPβ1 interaction and the subsequent modification of KAPβ1 remains to be characterized.
Additionally, two stress sensors in the unfolded protein response (UPR), namely PERK (protein kinase RNA-like ER kinase) and IRE1α (inositol-requiring transmembrane kinase and endonuclease 1α), were identified as interaction partners and substrates of ARTD15 (Ref. 55), suggesting that ARTD15 functions in the UPR in the ER57. A key regulator of these stress sensors is BiP (binding immunoglobulin protein; also known as GRP78). It interacts with PERK and IRE1α and thus inhibits them and, moreover, recognizes misfolded proteins. ARTD15 promotes the dissociation of BiP from PERK and IRE1α, possibly by binding to their peptide-binding pockets in the ER lumen, thereby displacing BiP. This, together with the stimulation of PERK kinase activity and both the kinase and endonuclease activities of IRE1α by MARylation, activates these ER stress sensors during the UPR (Fig. 1d). This is underlined by the fact that, upon knockdown of ARTD15, PERK and IRE1α cannot be activated, leading to cell death when the UPR is induced55. How ARTD15 is regulated is unclear, however, on the basis of structural studies, it was proposed that an α-helical domain, which is packed against the transferase domain of ARTD15, might be important in controlling its activity58.
Interestingly, in most of these reports, the observed phenomena rely not only on the catalytic activity of these different ARTD enzymes, but also on additional domains within these enzymes, such as ubiquitin-interaction motifs and zinc-finger domains, supporting the notion that these domains participate in substrate and pathway selection. These reports on mono-ADP-ribosyltransferases highlight the important roles of MARylation in diverse cellular processes such as NF-κB signalling, gene transcription and the UPR, although the exact underlying mechanisms need to be investigated in more detail.
Macrodomain-containing proteins
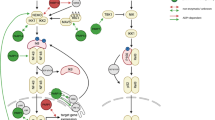
Although the first macrodomain was identified more than 20 years ago, the tight connection between these domains and ADP-ribosylation is only just being realized. Eleven human macrodomain-containing proteins have now been identified (Table 2). As discussed below, some of these macrodomains regulate PARylation, whereas it is emerging that others control MARylation (Fig. 2A).
A | Schematic summary of the emerging functions of macrodomains as readers and erasers of mono(ADP-ribosyl)ation (MARylation). Protein substrates are MARylated by ADP-ribosyltransferases (ARTs) ('ART writers'); this ADP-ribose moiety can be read by certain macrodomains (macroreader) or erased by other macrodomains (macroeraser). B | Macrodomains as readers of ADP-ribosylation. Certain macrodomains interact with poly(ADP-ribose) (PAR), including the macrodomains of DNA repair factors (Ba). Other macrodomains, for example macrodomain 3 of ARTD8, recognize ARTD10-mediated MARylation (Bb). We propose that this will serve either to recruit other proteins or to induce further signalling through intrinsic domains, such as the ART domain in ARTD8. C | Macrodomains as erasers of ADP-ribosylation. Glycogen synthase kinase 3β (GSK3β) is MARylated by ARTD10, leading to a decrease in its kinase activity. MacroD2 removes the MARylation, which reverts the inhibitory effect and increases GSK3β kinase activity (Ca). In addition to removal of MARylation, C6orf130 may also release entire PAR chains from PARylated proteins. This activity needs to be characterized in more detail (Cb). Poly(ADP-ribose) glycohydrolase (PARG) degrades PAR chains on ARTD1 but leaves the last ADP-ribose attached to the protein (Cc). This MAR moiety then potentially serves as a scaffold to recruit MAR readers to initiate further responses. Alternatively, ARTD1 MARylation serves as a substrate of MAR hydrolases, including macroD2, which revert the protein into its native state. Relevant proteins are depicted or examples are given if the activity demonstrated is shared by several macrodomains. Of note, these activities are not mutually exclusive as several macrodomains exhibit more than one function (summarized in Table 2).
The discovery of macrodomain-containing proteins. The macrodomain, which is a globular module, was identified as a domain of the histone variant macroH2A, and at the time it was of unknown function17. Macrodomains are not only found in vertebrates but also in many bacteria, archaea, viruses and plants, suggesting their evolutionary conservation59,60,61. Their structural characteristics are described in more detail in Box 1. Later, it became clear that macrodomains can interact with PARylated proteins and serve as binding modules for free ADP-ribose, as demonstrated first for the macrodomain of the Archaeoglobus fulgidus protein Af1521 and the macrodomains of ARTD9 (Ref. 62). Moreover, a catalytic activity has been assigned to some macrodomain-containing proteins, namely hydrolysis of O-acetyl-ADP-ribose63,64, which is a metabolite from the sirtuin-mediated NAD+-dependent deacetylation reaction65. Additionally, some macrodomains possess phosphatase activity towards ADP-ribose-1′′-phosphate, which is a product of tRNA splicing60,66. This phosphatase activity is widespread and found in macrodomain-containing proteins in viruses, bacteria and eukaryotes62,66,67,68,69. However, the observed reaction velocity is ranked as low throughout these studies67,68, raising the question of how physiologically relevant this activity is67. In accordance, this macrodomain phosphatase activity does not seem to be essential for the replication of human coronavirus in tissue culture70. Thus, the relevance of this activity remains to be investigated. Currently, the roles of macrodomain-containing proteins have been studied most extensively for PARylation.
Macrodomains read PARylation. Recognition of PAR is a well-established feature for certain macrodomains. In particular, PARylation in response to DNA damage is sensed by several macrodomains, which serve as recruitment modules for proteins involved in DNA repair (Fig. 2Ba). Both the histone variant macroH2A1.1 (Ref. 26) and the chromatin remodeller ALC1 are recruited in this manner and cause chromatin rearrangement and nucleosome sliding to facilitate DNA repair20,25,71. ARTD9 was identified as a risk factor in diffuse large B cell lymphoma72 as it is overexpressed in this disease, and it was shown to interact with the E3 ligase BBAP (B lymphoma- and BAL-associated protein; also known as DTX3L)73, although the exact function of the ARTD9–BBAP interaction remained undefined. However, it was recently suggested that ARTD9, which possesses two macrodomains, is recruited to ARTD1-generated PAR at DNA damage sites via its macrodomain 2, and that this is followed by BBAP-dependent local ubiquitylation and the subsequent recruitment of BRCA1 (breast cancer 1) and 53BP1 (p53-binding protein 1)74. ARTD9 does not seem to have any catalytic activity75. As the macrodomain 2, but not the macrodomain 1, can localize ARTD9 to PAR chains62,74, the function of macrodomain 1 is unclear. Additionally, the recruitment of both macroD1 and macroD2 to PAR synthesized by ARTD1 at damaged sites was observed in cells, but the consequences of this remain to be investigated26. This finding contradicts an earlier report that suggested that only macroD1 can efficiently bind PAR66. A possible explanation for this paradox might be the unnoticed presence of MAR at sites of DNA damage, to which macroD2 might be recruited. More recently, two reports have documented that macroD2 and C6orf130 (both are O-acetyl-ADP-ribose deacetylases), which also contain a macrodomain (Table 2), localize to DNA damage sites13,76. Because olaparib, an ARTD1 inhibitor, blocks this interaction, the initial activation of ARTD1 and PAR formation seem to be important for this. Time course experiments revealed the delayed binding of macroD2 compared with macroH2A1.1 to PAR at DNA damage sites, suggesting that macroD2 may bind to short PAR chains or to MARylated proteins upon PAR catabolism by PARG. This explains the recruitment of macroD2 to sites of DNA damage76. A catalytically inactive mutant of PARG has been used to selectively precipitate PARylated proteins from cells, and proteins with various functions, including multiple proteins associated with DNA damage, were identified77. Several macrodomain-containing proteins are thus closely linked with the ARTD family, PARylation and DNA repair. They have also been associated with different diseases (Box 2). New evidence points to a role for macrodomain-containing proteins in MAR signalling as well.
Macrodomains and MARylation
In contrast to the four binding modules that have been characterized for PAR, such modules were not identified for MARylated proteins until very recently. Clearly, macrodomains specify one motif that can read and erase PAR. It was therefore a logical step to address whether macrodomains are also involved in reading and processing MARylation (Fig. 2A).
Macrodomains as readers of MARylation. The first hint that macrodomain-containing proteins also recognize MARylated proteins came from pull-down experiment in which the macrodomain of the Af1521 protein from A. fulgidus was used to selectively precipitate ADP-ribosylated proteins from cell lysates78. It was postulated that the identified proteins are MARylated, although it was unclear whether this was indeed the case in these experiments. Moreover, because the macrodomain used was of bacterial origin and also capable of binding PARylated proteins62, the question remained whether endogenous human macrodomain-containing proteins exist that can bind to MARylated proteins in cells.
Recently, the crystal structures of the three ARTD8 macrodomains were solved, and the ADP-ribose-binding characteristics were determined79. Macrodomain 2 and macrodomain 3 from ARTD8 bind MARylated proteins with high-affinity, including automodified ARTD10 and MARylated proteins modified by ARTD10, such as the small GTPase RAN (Fig. 2Bb). These interactions are specific as neither macrodomain 2 nor macrodomain 3 of ARTD8 interacts with PARylated substrates. By contrast, macrodomain 1 of ARTD8 does not interact with MARylated or PARylated proteins79. Furthermore, the macrodomains of ARTD8 precipitated the ARTD10 substrate NEMO from cells co-expressing ARTD10 (Ref. 44), indicating that these macrodomains can be used as tools to confirm MARylation that is mediated by the ARTD family in cells. The ability of macrodomains to recognize MAR attached to proteins is not a general feature of macrodomains, unlike the interaction of macrodomains with free ADP-ribose. Instead, the binding capacity seems to be restricted to specific macrodomains as the macrodomain of macroH2A1.1 is unable to bind MARylated ARTD10 (Ref. 79).
These findings raise important questions. For example, what determines substrate-binding specificity? Because a single ADP-ribose fits into a macrodomain cleft it is tempting to speculate that macrodomains specifically binding to MARylated proteins, but not to PAR, also interact with part of the underlying protein, as is the case, for example, for bromodomains that interact with acetylated proteins80. Moreover, the backbone sequence around the MARylation site may determine the substrate specificity of macrodomains, with macrodomains perhaps even recognizing a sequence or structure that is similar to the modifying ARTD enzyme. In order to expand on these initial findings and to clarify the open questions, future research is required to investigate which features of the macrodomain-containing proteins mediate binding to MARylated proteins.
Macrodomains as erasers of MARylation. For a long time it was unclear which enzymes remove MAR from target proteins. PARG (which contains a macrodomain fold) and the structurally unrelated glycohydrolase ARH3 both breakdown PAR chains21,39, but neither can remove the final ADP-ribose moiety from the protein. Additionally, ARH3 was reported to hydrolyse O-acetyl-ADP-ribose81. ARH1 removes MAR from target proteins, but is ADP-ribosyl-Arg specific82. It has been proposed that intracellular ARTD enzymes modify either Glu or Lys, but not Arg, which makes it unlikely that ARH1 hydrolyses MAR transferred by mono-ADP-ribosyltransferases.
The gap in our knowledge on the reversibility of MARylation seems to be filled by the characterization of the O-acetyl-ADP-ribose deacetylases macroD1, macroD2 and C6orf13063,64 as hydrolases of MARylated proteins in three recent reports13,40,76. In all three studies, ARTD10-mediated MARylation could be reversed in vitro by these three enzymes dependent on their macrodomains. Moreover, ARTD10 MARylated histones and GSK3β were also substrates of macroD2, and the removal of MAR from GSK3β was sufficient to reactivate its kinase activity (Fig. 2Ca). It is noteworthy that C6orf130 may also be capable of releasing complete PAR chains from PARylated proteins13 (Fig. 2Cb). It has been suggested that free PAR chains serve as signalling molecules, leading to cell death83. These hydrolases seem to be functionally relevant as overexpression of macroD2 in cells increases GSK3β activity40, probably by removing the MAR that has been attached to it by ARTD10 (Ref. 47) (Fig. 2Ca). Moreover, PARylated ARTD1, when incubated with PARG, can be further de-ADP-ribosylated by macroD2, supporting the notion that PARG generates MARylated ARTD1 (Refs 13,40,76) (Fig. 2Cc). Conversely, the MAR-binding macrodomain 2 of ARTD8 does not display hydrolytic activity towards MARylated ARTD10 or its substrates40. These different functionalities imply that, despite their structural similarities, key residues are present in different macrodomains that mediate these activities. The biochemical studies suggest that macroD1 and macroD2 hydrolyse ADP-ribosylated Glu residues with ADP-ribose being linked to the γ-carboxylate of Glu through C1′′ of the distal ribose, requiring an H2O for the nucleophile attack40,76. These findings support the dissociative SN1-type mechanism of substrate-assisted catalysis proposed for ARTD10 (Ref. 8), a process in which the negatively charged γ-carboxylate of Glu participtates first in the destabilization of the glycosidic bond between nicotinamide and the ribose, followed by the release of nictonamide and the nucleophilic attack of the γ-carboxylate on the ribose, that is, on the oxocarbenium ion generated at C1′′ during the transition state. Interestingly, the role of Glu residues as substrates for ADP-ribosylation is documented by a case study of a patient suffering from a lysosomal storage disease. It was found that glutamyl ribose 5-phosphate accumulated in the lysosomes of this patient84,85, arguing for the relevance of enzymes that remove MAR from Glu residues. This is strongly supported by the identification of mutations in the gene encoding C6orf130 that are linked to a severe neurodegenerative phenotype13. This dramatic consequence of the lack of C6orf130 activity suggests that macroD1, macroD2 and C6orf130 have non-redundant functions.
These initial findings reveal the existence of MAR hydrolases that oppose ARTD-mediated mono-ADP-ribosyltransferase activities and establish MARylation as a reversible PTM (Fig. 2A). Moreover, the findings that mutations in the gene encoding C6orf130 and the accumulation of glutamyl ribose 5-phosphate are associated with neurodegeneration highlights the importance of controlling MARylation13,84,85. Nevertheless, more research is needed to dissect the precise function of MAR hydrolases. For instance, it should be addressed whether these enzymes are specific for distinct substrates by recognizing the peptide backbone underlying the MARylation site, or whether they generally remove MARylation and are regulated, for example, in a spatio-temporal manner.
Perspectives
The emerging functions of MARylation highlight the potential key roles of this PTM (Fig. 1). The thus far uncovered processes involving MARylation imply that this modification has a role in diverse signalling networks, including the UPR and, through its ability to influence NF-κB signalling, inflammatory processes. However, this is probably only the 'tip of the iceberg', and more work is needed to dissect the consequences of MARylation for target proteins and their associated pathways, as well as to further characterize the enzymes responsible for adding and removing this modification. The identification of macrodomains as binding modules suggests that there are mechanisms that can sense MARylation and likely respond to it through the associated proteins. Furthermore, these reports provide the first proof that endogenous intracellular MARylation actually exists. The characterization of macrodomain-containing proteins as hydrolases of MARylated proteins indicates that this modification is reversible, like other PTMs, and can thus form a transient signal in response to distinct stimuli (Fig. 2). To fully apprehend the function of MARylation, better tools and reliable methods must be developed (for example, antibodies) to be able to measure MARylation in cells and to map modification sites within target proteins.
Macrodomain-containing proteins also require further investigation. Currently, there is only a limited number of proteins with identified macrodomains. Because the macrodomain fold in PARG could not be deduced from sequence alignments alone, more macrodomain-containing proteins might be discovered purely based on their structure. The known macrodomain-containing proteins (Table 2) should be studied in more detail to determine whether they bind poly(ADP-ribose) or mono(ADP-ribose) and what activity they have. Moreover, it is not entirely clear why some macrodomains only bind PAR chains and others only MAR, or why some have catalytic activities whereas others do not (apart from PARG in which the catalytic loop enables PARG to breakdown PAR chains). The crucial residues for these different functions need to be investigated in more detail. Future research should thus be directed at improved understanding of the binding and catalytic mechanisms of macrodomains, to be able to determine their roles in the regulation of MARylation. One study indicates that ARTD10 and ARTD8 together modify roughly 200 proteins in vitro47. This is probably an underestimation, as the screen did not sample all proteins, but not all the in vitro targets may be true substrates in cells. Despite this uncertainty, assuming that each of the nine proposed mono-ADP-ribosyltransferases possesses 100 substrates and considering that the removal of PAR from substrates by PARG and ARH3 potentially results in MARylated substrates, it is possible that more than 1000 MARylated proteins exist. Thus, it is likely that in addition to macrodomains further domains exist that read and erase MARylation. It will be interesting to follow how this research area will develop, which substrates and pathways are sensitive to MARylation and how this PTM is interpreted and regulated in cells.
Change history
28 June 2013
On page 443 of this article, the incorrect references were cited at the end of the following sentence: "Four years ago, macrodomains were shown to be able to bind PAR that has been synthesized in response to DNA damage18,19,20." The authors meant to refer to references 20, 25 and 26 at the end of this sentence. This has been corrected online. The authors apologize to the authors of references 25 and 26 for this mistake and for any confusion caused to readers.
References
Holbourn, K. P., Shone, C. C. & Acharya, K. R. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 273, 4579–4593 (2006).
Corda, D. & Di Girolamo, M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 22, 1953–1958 (2003).
Yates, S. P., Jorgensen, R., Andersen, G. R. & Merrill, A. R. Stealth and mimicry by deadly bacterial toxins. Trends Biochem. Sci. 31, 123–133 (2006).
Otto, H. et al. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 6, 139 (2005).
Hottiger, M. O., Hassa, P. O., Luscher, B., Schuler, H. & Koch-Nolte, F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 35, 208–219 (2010).
Schreiber, V., Dantzer, F., Ame, J. C. & de Murcia, G. Poly(ADP-ribose): novel functions for an old molecule. Nature Rev. Mol. Cell Biol. 7, 517–528 (2006).
Gibson, B. A. & Kraus, W. L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature Rev. Mol. Cell Biol. 13, 411–424 (2012).
Kleine, H. et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell 32, 57–69 (2008).
Altmeyer, M., Messner, S., Hassa, P. O., Fey, M. & Hottiger, M. O. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 37, 3723–3738 (2009).
Haenni, S. S. et al. Identification of lysines 36 and 37 of PARP-2 as targets for acetylation and auto-ADP-ribosylation. Int. J. Biochem. Cell Biol. 40, 2274–2283 (2008).
Messner, S. et al. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 38, 6350–6362 (2010).
Chapman, J. D., Gagne, J. P., Poirier, G. G. & Goodlett, D. R. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 12, 1868–1880 (2013).
Sharifi, R. et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 32, 1225–1237 (2013).
Tao, Z., Gao, P. & Liu, H. W. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J. Am. Chem. Soc. 131, 14258–14260 (2009).
Kalisch, T., Ame, J. C., Dantzer, F. & Schreiber, V. New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem. Sci. 37, 381–390 (2012).
Zaja, R., Mikoc, A., Barkauskaite, E. & Ahel, I. Molecular insights into poly(ADP-ribose) recognition and processing. Biomolecules 3, 1–17 (2012).
Pehrson, J. R. & Fried, V. A. MacroH2A, a core histone containing a large nonhistone region. Science 257, 1398–1400 (1992).
Li, G. Y. et al. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc. Natl Acad. Sci. USA 107, 9129–9134 (2010).
Ahel, I. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008).
Ahel, D. et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325, 1240–1243 (2009).
Slade, D. et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477, 616–620 (2011).
Kim, I. K. et al. Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nature Struct. Mol. Biol. 19, 653–656 (2012).
Dunstan, M. S. et al. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nature Commun. 3, 878 (2012).
Satoh, M. S. & Lindahl, T. Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358 (1992).
Gottschalk, A. J. et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl Acad. Sci. USA 106, 13770–13774 (2009).
Timinszky, G. et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nature Struct. Mol. Biol. 16, 923–929 (2009).
Kleine, H. & Luscher, B. Learning how to read ADP-ribosylation. Cell 139, 17–19 (2009).
Huambachano, O., Herrera, F., Rancourt, A. & Satoh, M. S. Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J. Biol. Chem. 286, 7149–7160 (2011).
Hsiao, S. J. & Smith, S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90, 83–92 (2008).
Aravind, L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26, 273–275 (2001).
Callow, M. G. et al. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PLoS ONE 6, e22595 (2011).
Zhang, Y. et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nature Cell Biol. 13, 623–629 (2011).
MacDonald, B. T., Tamai, K. & He, X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 (2009).
Pleschke, J. M., Kleczkowska, H. E., Strohm, M. & Althaus, F. R. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275, 40974–40980 (2000).
Gagne, J. P. et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 36, 6959–6976 (2008).
Krietsch, J. et al. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Aspects Med. 23 Dec 2012 (10.1016/j.mam.2012.12.005).
Tucker, J. A. et al. Structures of the human poly (ADP-ribose) glycohydrolase catalytic domain confirm catalytic mechanism and explain inhibition by ADP-HPD derivatives. PLoS ONE 7, e50889 (2012).
Oka, S., Kato, J. & Moss, J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 281, 705–713 (2006).
Mueller-Dieckmann, C. et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl Acad. Sci. USA 103, 15026–15031 (2006).
Rosenthal, F. et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nature Struct. Mol. Biol. 20, 502–507 (2013).
Hottiger, M. O. et al. Progress in the function and regulation of ADP-ribosylation. Sci. Signal. 4, mr5 (2011).
Yu, M. et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene 24, 1982–1993 (2005).
Kleine, H. et al. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun. Signal. 10, 28 (2012).
Verheugd, P. et al. Regulation of NF-κB signaling by the mono-ADP-ribosyltransferase ARTD10. Nature Commun. 4, 1683 (2013).
Chen, Z. J. Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 246, 95–106 (2012).
Herzog, N. et al. Caspase-dependent cleavage of the mono-ADP-ribosyltransferase ARTD10 interferes with its pro-apoptotic function. FEBS J. 280, 13 (2013).
Feijs, K. L. et al. ARTD10 substrate identification on protein microarrays: regulation of GSK3β by mono-ADP-ribosylation. Cell Commun. Signal. 11, 5 (2013).
Wu, D. & Pan, W. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem. Sci. 35, 161–168 (2010).
Goenka, S. & Boothby, M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6 (CoaSt6), a transcriptional cofactor. Proc. Natl Acad. Sci. USA 103, 4210–4215 (2006).
Mehrotra, P. et al. PARP-14 functions as a transcriptional switch for Stat6-dependent gene activation. J. Biol. Chem. 286, 1767–1776 (2011).
Goenka, S., Cho, S. H. & Boothby, M. Collaborator of Stat6 (CoaSt6)-associated poly(ADP-ribose) polymerase activity modulates Stat6-dependent gene transcription. J. Biol. Chem. 282, 18732–18739 (2007).
Yang, J. et al. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 21, 4950–4958 (2002).
Mehrotra, P. et al. Poly (ADP-ribose) polymerase 14 and its enzyme activity regulates TH2 differentiation and allergic airway disease. J. Allergy Clin. Immunol. 131, 521–531.e12 (2013).
Macpherson, L. et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase (TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of aryl hydrocarbon receptor transactivation. Nucleic Acids Res. 41, 1604–1621 (2012).
Jwa, M. & Chang, P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nature Cell Biol. 14, 1223–1230 (2012).
Di Paola, S., Micaroni, M., Di Tullio, G., Buccione, R. & Di Girolamo, M. PARP16/ARTD15 is a novel endoplasmic-reticulum-associated mono-ADP-ribosyltransferase that interacts with, and modifies karyopherin-ss1. PLoS ONE 7, e37352 (2012).
Hetz, C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nature Rev. Mol. Cell Biol. 13, 89–102 (2012).
Karlberg, T., Thorsell, A. G., Kallas, A. & Schuler, H. Crystal structure of human ADP-ribose transferase ARTD15/PARP16 reveals a novel putative regulatory domain. J. Biol. Chem. 287, 24077–24081 (2012).
Anantharaman, V., Koonin, E. V. & Aravind, L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 30, 1427–1464 (2002).
Till, S. & Ladurner, A. G. Sensing NAD metabolites through macro domains. Front. Biosci. 14, 3246–3258 (2009).
Han, W., Li, X. & Fu, X. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat. Res. 727, 86–103 (2011).
Karras, G. I. et al. The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005).
Chen, D. et al. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 286, 13261–13271 (2011).
Peterson, F. C. et al. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J. Biol. Chem. 286, 35955–35965 (2011).
Tong, L. & Denu, J. M. Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochim. Biophys. Acta 1804, 1617–1625 (2010).
Neuvonen, M. & Ahola, T. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 385, 212–225 (2009).
Egloff, M. P. et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 80, 8493–8502 (2006).
Saikatendu, K. S. et al. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13, 1665–1675 (2005).
Shull, N. P., Spinelli, S. L. & Phizicky, E. M. A highly specific phosphatase that acts on ADP-ribose 1′′-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 33, 650–660 (2005).
Putics, A., Filipowicz, W., Hall, J., Gorbalenya, A. E. & Ziebuhr, J. ADP-ribose-1”-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79, 12721–12731 (2005).
Pines, A. et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J. Cell Biol. 199, 235–249 (2012).
Aguiar, R. C. et al. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood 96, 4328–4334 (2000).
Takeyama, K. et al. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem. 278, 21930–21937 (2003).
Yan, Q. et al. BAL1 and its partner E3 ligase, BBAP, link PARP activation, ubiquitylation and double-strand DNA repair independent of ATM, MDC1 and RNF8. Mol Cell. Biol. 33, 845–857 (2013).
Aguiar, R. C., Takeyama, K., He, C., Kreinbrink, K. & Shipp, M. A. B-aggressive lymphoma family proteins have unique domains that modulate transcription and exhibit poly(ADP-ribose) polymerase activity. J. Biol. Chem. 280, 33756–33765 (2005).
Jankevicius, G. et al. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nature Struct. Mol. Biol. 20, 508–514 (2013).
Gagne, J. P. et al. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 40, 7788–7805 (2012).
Dani, N. et al. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc. Natl Acad. Sci. USA 106, 4243–4248 (2009).
Forst, A. H. et al. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure 21, 462–475 (2013).
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012).
Ono, T., Kasamatsu, A., Oka, S. & Moss, J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc. Natl Acad. Sci. USA 103, 16687–16691 (2006).
Moss, J., Oppenheimer, N. J., West, R. E. Jr & Stanley, S. J. Amino acid specific ADP-ribosylation: substrate specificity of an ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry 25, 5408–5414 (1986).
Wang, Y. et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Sci. Signal. 4, ra20 (2011).
Williams, J. C., Chambers, J. P. & Liehr, J. G. Glutamyl ribose 5-phosphate storage disease. A hereditary defect in the degradation of poly(ADP-ribosylated) proteins. J. Biol. Chem. 259, 1037–1042 (1984).
Williams, J. C. et al. Progressive neurologic deterioration and renal failure due to storage of glutamyl ribose-5-phosphate. N. Engl. J. Med. 311, 152–155 (1984).
Allen, M. D., Buckle, A. M., Cordell, S. C., Lowe, J. & Bycroft, M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330, 503–511 (2003).
Kustatscher, G., Hothorn, M., Pugieux, C., Scheffzek, K. & Ladurner, A. G. Splicing regulates NAD metabolite binding to histone macroH2A. Nature Struct. Mol. Biol. 12, 624–625 (2005).
Patel, C. N., Koh, D. W., Jacobson, M. K. & Oliveira, M. A. Identification of three critical acidic residues of poly(ADP-ribose) glycohydrolase involved in catalysis: determining the PARG catalytic domain. Biochem. J. 388, 493–500 (2005).
Cho, S. H. et al. PARP-14, a member of the B aggressive lymphoma family, transduces survival signals in primary B cells. Blood 113, 2416–2425 (2009).
Ma, N. F. et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology 47, 503–510 (2008).
Brockschmidt, A. et al. CHD1L: a new candidate gene for congenital anomalies of the kidneys and urinary tract (CAKUT). Nephrol. Dial. Transplant. 27, 2355–2364 (2012).
Kapoor, A. et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 468, 1105–1109 (2010).
Sporn, J. C. et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 28, 3423–3428 (2009).
Park, E. & Griffin, D. E. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology 388, 305–314 (2009).
Chaurushiya, M. S. & Weitzman, M. D. Viral manipulation of DNA repair and cell cycle checkpoints. DNA Repair (Amst.) 8, 1166–1176 (2009).
Haince, J. F., Ouellet, M. E., McDonald, D., Hendzel, M. J. & Poirier, G. G. Dynamic relocation of poly(ADP-ribose) glycohydrolase isoforms during radiation-induced DNA damage. Biochim. Biophys. Acta 1763, 226–237 (2006).
Acknowledgements
The authors thank their colleagues for insightful discussions and A. Ladurner and I. Ahel for providing manuscripts prior to publication. They apologize to researchers whose work could not be included owing to space restrictions. The work in the author's laboratory was supported by the START program of the Medical School of the Rheinisch-Westfaelische Technische Hochschule (RWTH) Aachen University and by the Deutsche Forschungsgemeinschaft DFG (LU466/15-1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Feijs, K., Forst, A., Verheugd, P. et al. Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat Rev Mol Cell Biol 14, 443–451 (2013). https://doi.org/10.1038/nrm3601
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm3601
This article is cited by
-
Mono-ADP-ribosylation by PARP10 inhibits Chikungunya virus nsP2 proteolytic activity and viral replication
Cellular and Molecular Life Sciences (2023)
-
Intracellular mono-ADP-ribosyltransferases at the host–virus interphase
Cellular and Molecular Life Sciences (2022)
-
Fundamental mechanisms of the stem cell regulation in land plants: lesson from shoot apical cells in bryophytes
Plant Molecular Biology (2021)
-
A mycorrhizae-like gene regulates stem cell and gametophore development in mosses
Nature Communications (2020)
-
Engineering Af1521 improves ADP-ribose binding and identification of ADP-ribosylated proteins
Nature Communications (2020)