Key Points
-
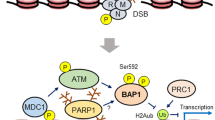
The breast and ovarian cancer susceptibility type 1 protein (BRCA1) exists as a heterodimer with BRCA1-associated RING domain protein 1 (BARD1) and forms at least three separate macromolecular protein complexes in vivo through abraxas (also known as CCDC98 and FAM175A), BRCA1-interacting protein carboxy-terminal helicase 1 (BACH1; also known as FANCJ and BRIP1) and CtBP-interacting protein (CtIP; also known as RBBP8), each of which regulates a specific set of responses following genotoxic stress.
-
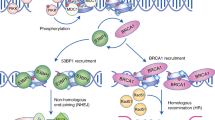
BRCA1 promotes optimal homologous recombination (HR)-mediated DNA repair by orchestrating various steps of the reaction: checkpoint activation, DNA end resection and RAD51 assimilation.
-
Partner and localizer of BRCA2 (PALB2; also known as FANCN) bridges the association between BRCA1 and BRCA2 (also known as FANCD1) and links BRCA1 directly to HR-mediated DNA repair.
-
An emerging ubiquitylation-dependent cascade, involving RING finger protein 8 (RNF8)-, RNF168- or ubiquitin-conjugating enzyme 13 (UBC13; also known as UBE2N)-catalysed ubiquitin chains and their recognition by the ubiquitin-interacting motif-containing receptor-associated protein 80 (RAP80; also known as UIMC1), governs BRCA1 localization in the vicinity of double-stranded DNA breaks.
-
BRCA1 seems to participate in a subset of DNA cross link repair pathways in concert with the BRCA-related Fanconi anaemia components, PALB2, BACH1 and BRCA2.
Abstract
The breast and ovarian cancer type 1 susceptibility protein (BRCA1) has pivotal roles in the maintenance of genome stability. Studies support that BRCA1 exerts its tumour suppression function primarily through its involvement in cell cycle checkpoint control and DNA damage repair. In addition, recent proteomic and genetic studies have revealed the presence of distinct BRCA1 complexes in vivo, each of which governs a specific cellular response to DNA damage. Thus, BRCA1 is emerging as the master regulator of the genome through its ability to execute and coordinate various aspects of the DNA damage response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005).
Gorgoulis, V. G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005).
Miki, Y. et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266, 66–71 (1994).
Manke, I. A., Lowery, D. M., Nguyen, A. & Yaffe, M. B. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302, 636–639 (2003). Together with reference 6, these studies laid the groundwork for how the BRCA1 BRCT domains interact with various binding partners.
Yu, X., Chini, C. C., He, M., Mer, G. & Chen, J. The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 (2003).
Meza, J. E., Brzovic, P. S., King, M. C. & Klevit, R. E. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J. Biol. Chem. 274, 5659–5665 (1999).
Ghimenti, C. et al. Germline mutations of the BRCA1-associated ring domain (BARD1) gene in breast and breast/ovarian families negative for BRCA1 and BRCA2 alterations. Genes Chromosomes Cancer 33, 235–242 (2002).
Thai, T. H. et al. Mutations in the BRCA1-associated RING domain (BARD1) gene in primary breast, ovarian and uterine cancers. Hum. Mol. Genet. 7, 195–202 (1998).
Capasso, M. et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nature Genet. 41, 718–723 (2009).
Westermark, U. K. et al. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol. Cell. Biol. 23, 7926–7936 (2003).
Brzovic, P. S., Rajagopal, P., Hoyt, D. W., King, M. C. & Klevit, R. E. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nature Struct. Biol. 8, 833–837 (2001).
Christensen, D. E., Brzovic, P. S. & Klevit, R. E. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nature Struct. Mol. Biol. 14, 941–948 (2007). Suggests that BRCA1 may promote the formation of distinct ubiquitin chains through selective interaction with E2 enzymes.
Brzovic, P. S., Lissounov, A., Christensen, D. E., Hoyt, D. W. & Klevit, R. E. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 (2006).
Parvin, J. D. The BRCA1-dependent ubiquitin ligase, γ-tubulin, and centrosomes. Environ. Mol. Mutagen. 50, 649–653 (2009).
Heine, G. F. & Parvin, J. D. BRCA1 control of steroid receptor ubiquitination. Sci. STKE. 2007, pe34 (2007).
Starita, L. M. & Parvin, J. D. Substrates of the BRCA1-dependent ubiquitin ligase. Cancer Biol. Ther. 5, 137–141 (2006).
Eakin, C. M., Maccoss, M. J., Finney, G. L. & Klevit, R. E. Estrogen receptor α is a putative substrate for the BRCA1 ubiquitin ligase. Proc. Natl Acad. Sci. USA 104, 5794–5799 (2007).
Anderson, S. F., Schlegel, B. P., Nakajima, T., Wolpin, E. S. & Parvin, J. D. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nature Genet. 19, 254–256 (1998).
Scully, R. et al. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl Acad. Sci. USA 94, 5605–5610 (1997).
Bochar, D. A. et al. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102, 257–265 (2000).
Yarden, R. I. & Brody, L. C. BRCA1 interacts with components of the histone deacetylase complex. Proc. Natl Acad. Sci. USA 96, 4983–4988 (1999).
Scully, R. et al. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88, 265–275 (1997). First evidence for a role of BRCA1 in HR-based DNA repair
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 (1998).
Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Celeste, A. et al. Genomic instability in mice lacking histone H2AX. Science 296, 922–927 (2002).
Stucki, M. et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 (2005).
Lou, Z., Minter-Dykhouse, K., Wu, X. & Chen, J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421, 957–961 (2003).
Stewart, G. S., Wang, B., Bignell, C. R., Taylor, A. M. & Elledge, S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966 (2003).
Lou, Z., Chini, C. C., Minter-Dykhouse, K. & Chen, J. Mediator of DNA damage checkpoint protein 1 regulates BRCA1 localization and phosphorylation in DNA damage checkpoint control. J. Biol. Chem. 278, 13599–13602 (2003).
Goldberg, M. et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421, 952–956 (2003).
Xu, X. & Stern, D. F. NFBD1/MDC1 regulates ionizing radiation-induced focus formation by DNA checkpoint signaling and repair factors. Faseb J. 17, 1842–1848 (2003).
Xu, X. & Stern, D. F. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J. Biol. Chem. 278, 8795–8803 (2003).
Yu, X., Wu, L. C., Bowcock, A. M., Aronheim, A. & Baer, R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273, 25388–25392 (1998).
Cantor, S. B. et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105, 149–160 (2001).
Wang, B. et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 (2007).
Kim, H., Huang, J. & Chen, J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nature Struct. Mol. Biol. 14, 710–715 (2007).
Liu, Z., Wu, J. & Yu, X. CCDC98 targets BRCA1 to DNA damage sites. Nature Struct. Mol. Biol. 14, 716–720 (2007).
Yan, J. et al. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 67, 6647–6656 (2007).
Kim, H., Chen, J. & Yu, X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007). References 36–41 reveal mechanistically how BRCA1 is targeted to sites of DNA breaks.
Wang, B. & Elledge, S. J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl Acad. Sci. USA 104, 20759–20763 (2007).
Kolas, N. K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007).
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Hofmann, R. M. & Pickart, C. M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999).
Lorick, K. L. et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl Acad. Sci. USA 96, 11364–11369 (1999).
Morris, J. R. & Solomon, E. BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum. Mol. Genet. 13, 807–817 (2004). Together with reference 56, these studies reveal that BRCA1-dependent non-canonical types of ubiquitin linkages are required for DNA repair.
Polanowska, J., Martin, J. S., Garcia-Muse, T., Petalcorin, M. I. & Boulton, S. J. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 25, 2178–2188 (2006).
Chen, A., Kleiman, F. E., Manley, J. L., Ouchi, T. & Pan, Z. Q. Autoubiquitination of the BRCA1•BARD1 RING ubiquitin ligase. J. Biol. Chem. 277, 22085–22092 (2002).
Zhao, G. Y. et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25, 663–675 (2007).
Yu, X. & Chen, J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24, 9478–9486 (2004).
Yu, X., Fu, S., Lai, M., Baer, R. & Chen, J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20, 1721–1726 (2006).
Nishikawa, H. et al. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 279, 3916–3924 (2004).
Xia, Y., Pao, G. M., Chen, H. W., Verma, I. M. & Hunter, T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 278, 5255–5263 (2003).
Foray, N. et al. A subset of ATM- and ATR-dependent phosphorylation events requires the BRCA1 protein. EMBO J. 22, 2860–2871 (2003).
Yarden, R. I., Pardo-Reoyo, S., Sgagias, M., Cowan, K. H. & Brody, L. C. BRCA1 regulates the G2/M checkpoint by activating CHK1 kinase upon DNA damage. Nature Genet. 30, 285–289 (2002). Identifies BRCA1 function in the G2–M checkpoint control through regulation of CHK1 phosphorylation
Wu, L. C. et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nature Genet. 14, 430–440 (1996).
Baer, R. & Ludwig, T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12, 86–91 (2002).
Starita, L. M. et al. BRCA1-dependent ubiquitination of γ-tubulin regulates centrosome number. Mol. Cell. Biol. 24, 8457–8466 (2004).
Hashizume, R. et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 276, 14537–14540 (2001).
Nishikawa, H. et al. BRCA1-associated protein 1 interferes with BRCA1/BARD1 RING heterodimer activity. Cancer Res. 69, 111–119 (2009).
Shakya, R. et al. The basal-like mammary carcinomas induced by BRCA1 or BARD1 inactivation implicate the BRCA1/BARD1 heterodimer in tumor suppression. Proc. Natl Acad. Sci. USA 105, 7040–7045 (2008).
Joukov, V., Chen, J., Fox, E. A., Green, J. B. & Livingston, D. M. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl Acad. Sci. USA 98, 12078–12083 (2001).
Greenberg, R. A. et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20, 34–46 (2006). A proposed model of how BRCA1 regulates diverse cellular processes in the DNA damage response.
Fernandez-Capetillo, O. et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nature Cell Biol. 4, 993–997 (2002).
Chen, X., Arciero, C. A., Wang, C., Broccoli, D. & Godwin, A. K. BRCC36 is essential for ionizing radiation-induced BRCA1 phosphorylation and nuclear foci formation. Cancer Res. 66, 5039–5046 (2006).
Dong, Y. et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell 12, 1087–1099, (2003).
Ambroggio, X. I., Rees, D. C. & Deshaies, R. J. JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2, E2 (2004).
Wang, B., Hurov, K., Hofmann, K. & Elledge, S. J. NBA1, a new player in the BRCA1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 23, 729–739 (2009).
Feng, L., Huang, J. & Chen, J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 23, 719–728 (2009).
Shao, G. et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23, 740–754 (2009).
Xu, B., O'Donnell, A. H., Kim, S. T. & Kastan, M. B. Phosphorylation of serine 1387 in BRCA1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res. 62, 4588–4591 (2002).
Xu, B., Kim, S. & Kastan, M. B. Involvement of BRCA1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21, 3445–3450 (2001).
Kumaraswamy, E. & Shiekhattar, R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol. Cell. Biol. 27, 6733–6741 (2007).
Makiniemi, M. et al. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276, 30399–30406 (2001).
Kim, J. E., McAvoy, S. A., Smith, D. I. & Chen, J. Human TopBP1 ensures genome integrity during normal S phase. Mol. Cell. Biol. 25, 10907–10915 (2005).
Van Hatten, R. A. et al. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159, 541–547 (2002).
Hashimoto, Y. & Takisawa, H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 22, 2526–2535 (2003).
Huertas, P. & Jackson, S. P. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284, 9558–9565 (2009).
Sartori, A. A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007).
Mimitou, E. P. & Symington, L. S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008).
Nimonkar, A. V., Ozsoy, A. Z., Genschel, J., Modrich, P. & Kowalczykowski, S. C. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl Acad. Sci. USA 105, 16906–16911 (2008).
Gravel, S., Chapman, J. R., Magill, C. & Jackson, S. P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22, 2767–2772 (2008).
Chen, L., Nievera, C. J., Lee, A. Y. & Wu, X. Cell cycle-dependent complex formation of BRCA1•CtIP•MRN is important for DNA double-strand break repair. J. Biol. Chem. 283, 7713–7720 (2008).
Schlegel, B. P., Jodelka, F. M. & Nunez, R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 66, 5181–5189 (2006).
Yun, M. H. & Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 (2009).
Zou, L. & Elledge, S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003).
Venere, M., Snyder, A., Zgheib, O. & Halazonetis, T. D. Phosphorylation of ATR-interacting protein on Ser239 mediates an interaction with breast-ovarian cancer susceptibility 1 and checkpoint function. Cancer Res. 67, 6100–6105 (2007). The first documented BRCA1 accumulation at two distinct micronuclear domains, which correspond to single-stranded and double-stranded DNA breaks.
Bekker-Jensen, S. et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195–206 (2006).
Celeste, A. et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nature Cell Biol. 5, 675–679 (2003).
Celeste, A. et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114, 371–383 (2003).
Lou, Z. et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21, 187–200 (2006).
Zhong, Q. et al. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science 285, 747–750 (1999).
Moynahan, M. E., Chiu, J. W., Koller, B. H. & Jasin, M. Brca1 controls homology-directed DNA repair. Mol. Cell 4, 511–518 (1999). The first functional evidence for a role of BRCA1 in HR repair.
Chen, J. et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol. Cell 2, 317–328 (1998).
Thorslund, T. & West, S. C. BRCA2: a universal recombinase regulator. Oncogene 26, 7720–7730 (2007).
Bhattacharyya, A., Ear, U. S., Koller, B. H., Weichselbaum, R. R. & Bishop, D. K. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275, 23899–23903 (2000).
Xia, B. et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729 (2006).
Erkko, H. et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature 446, 316–319 (2007).
Xia, B. et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nature Genet. 39, 159–161 (2007).
Reid, S. et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nature Genet. 39, 162–164 (2007).
Rahman, N. et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nature Genet. 39, 165–167 (2007).
Sy, S. M., Huen, M. S. & Chen, J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl Acad. Sci. USA 106, 7155–7160 (2009). Together with reference 107, these studies identify PALB2 as the link between BRCA1 and proteins involved in HR-mediated repair.
Zhang, F. et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 19, 524–529 (2009).
Zhang, J. et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol. Cell Biol. 24, 708–718 (2004).
Reid, L. J. et al. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl Acad. Sci. USA 105, 20876–20881 (2008).
Lou, Z., Minter-Dykhouse, K. & Chen, J. BRCA1 participates in DNA decatenation. Nature Struct. Mol. Biol. 12, 589–593 (2005).
Wang, W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nature Rev. Genet. 8, 735–748 (2007).
Kennedy, R. D. & D'Andrea, A. D. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19, 2925–2940 (2005).
Garcia-Higuera, I. et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7, 249–262 (2001).
Peng, M. et al. The FANCJ/MutLα interaction is required for correction of the cross-link response in FA-J. cells. EMBO J. 26, 3238–3249 (2007).
Bridge, W. L., Vandenberg, C. J., Franklin, R. J. & Hiom, K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nature Genet. 37, 953–957 (2005).
Niedernhofer, L. J., Lalai, A. S. & Hoeijmakers, J. H. Fanconi anemia (cross)linked to DNA repair. Cell 123, 1191–1198 (2005).
Shen, X. et al. Recruitment of Fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol. Cell 35, 716–723 (2009).
Cao, L. et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by BRCA1 deficiency. Mol. Cell 35, 534–541 (2009). In vivo evidence of an important link between BRCA1- and TP53BP1-dependent pathways.
Brazda, V., Jagelska, E. B., Liao, J. C. & Arrowsmith, C. H. The central region of BRCA1 binds preferentially to supercoiled DNA. J. Biomol. Struct. Dyn. 27, 97–104 (2009).
Shao, G. et al. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl Acad. Sci. USA 106, 3166–3171 (2009).
Bartek, J., Lukas, J. & Bartkova, J. DNA damage response as an anti-cancer barrier: damage threshold and the concept of 'conditional haploinsufficiency'. Cell Cycle 6, 2344–2347 (2007).
Bartek, J. & Lukas, J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19, 238–245 (2007).
Li, X. & Heyer, W. D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113 (2008).
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (CA089239, CA092312 and CA100109 to J.C.), the Startup Fund (Department of Anatomy, The University of Hong Kong to M.S.Y.H.) and Seed Funding for Basic Research (The University of Hong Kong to M.S.Y.H.). J.C is a recipient of an Era of Hope Scholar award from the Department of Defence and a member of the Mayo Clinic Breast SPORE program (P50 CA116201).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- E3 ubiquitin ligase
-
A protein or protein complex that covalently attaches ubiquitin moieties to its target protein by an isopeptide bond. E3 ubiquitin ligases usually provide the substrate specificity for a ubiquitylation reaction that involves an E1 ubiquitin-activating enzyme and an E2 ubiquitin-conjugating enzyme. Two major classes of E3 ubiquitin ligases have been defined based on their conserved HECT and RING domains.
- Homologous recombination
-
A DNA recombination pathway, which includes the repair of dsDNA breaks, that uses a homologous dsDNA molecule as a template for the repair of the broken DNA.
- E2 ubiquitin-conjugating enzyme
-
A protein that transfers the activated ubiquitin from the E1 ubiquitin-activating enzyme to an E3 ubiquitin ligase. E2 ubiquitin-conjugating enzymes determine ubiquitin chain specificities, and each E2 enzyme associates with several E3 ligases.
- Coiled-coil domain
-
A structural element that is important for mediating protein–protein interactions. The sequence of coiled-coil domains contains repetitive elements of seven apolar residues that form a heptad.
- Non-canonical ubiquitin chain
-
A diubiquitin or polyubiquitin chain comprising ubiquitin molecules that are conjugated by their Lys residues, other than Lys48. Some of these chains do not target proteins for proteosomal degradation.
- G2–M checkpoint
-
A DNA damage-induced transient cell cycle arrest at the G2–M border, usually associated with CHK1 phosphorylation and activation.
- Proteasome
-
A large protein complex that is responsible for breaking down polyubiquitylated proteins.
- G1–S checkpoint
-
A checkpoint that ensures growing conditions are optimal before cells are committed to one round of DNA replication and cell division.
- Intra-S phase checkpoint
-
ATM- and ATR-dependent transient inhibition of DNA replication in response to DNA damage. Defects in an ionising radiation-induced intra-S phase checkpoint cause radioresistant DNA synthesis.
- DEAH helicase family
-
A family of proteins that use ATP and unwind nucleic acids, which have a conserved DEAH box.
- Radial chromosome
-
An abnormal chromosome structure that results from pairing of homologous or non-homologous metaphase chromosomes. These structures are observed in chromosome spreads prepared from cells with an underlying chromosome instability, such as cells from patients with Fanconi anaemia, Bloom syndrome and ataxia telangiectasia.
- Gene conversion
-
A non-reciprocal recombination process that results in an alteration of the sequence of a gene to that of its homologue.
- Isogenic
-
Here, refers to a genotype that has been engineered to be identical to another genotype, with the exception of one or more mutations of interest.
- DNA decatenation
-
An ATP-dependent process for the resolution of replicated sister chromatids that requires topoisomerase II activity.
- Chromatid exchange
-
The physical exchange of genetic material between identical sister chromatids. This process can be enhanced by treatment with DNA damaging agents, such as the cross-linking agent mitomycin C.
Rights and permissions
About this article
Cite this article
Huen, M., Sy, S. & Chen, J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 11, 138–148 (2010). https://doi.org/10.1038/nrm2831
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm2831
This article is cited by
-
BRCA genes as candidates for colorectal cancer genetic testing panel: systematic review and meta-analysis
BMC Cancer (2023)
-
Integrative phosphoproteomics defines two biologically distinct groups of KMT2A rearranged acute myeloid leukaemia with different drug response phenotypes
Signal Transduction and Targeted Therapy (2023)
-
UBE2T regulates epithelial–mesenchymal transition through the PI3K-AKT pathway and plays a carcinogenic role in ovarian cancer
Journal of Ovarian Research (2022)
-
BRCA1 mutations in high-grade serous ovarian cancer are associated with proteomic changes in DNA repair, splicing, transcription regulation and signaling
Scientific Reports (2022)
-
HspBP1 is a dual function regulatory protein that controls both DNA repair and apoptosis in breast cancer cells
Cell Death & Disease (2022)