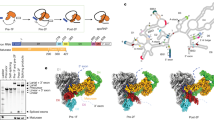
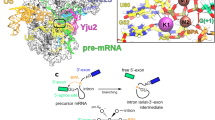
Key Points
-
Analysis of the recent atomic-resolution cryo-electron microscopy structures of the spliceosome reveals that during splicing all spliceosomal complexes share a rigid core of 20 RNAs and proteins.
-
The active site of the spliceosome is positioned in the catalytic cavity of pre-mRNA-splicing factor 8 (Prp8), is stabilized by surrounding proteins and remains static throughout the two steps of transesterification.
-
The catalytic metals in the active site are coordinated by U6 small nuclear RNA and the catalytic triplex.
-
The interconversion of the various spliceosomal complexes is driven by eight conserved, RNA-dependent ATPase/helicases.
-
The spliceosome is a protein-directed metalloribozyme.
Abstract
Precursor messenger RNA (pre-mRNA) splicing is an essential step in the flow of information from DNA to protein in all eukaryotes. Research over the past four decades has molecularly delineated the splicing pathway, including characterization of the detailed splicing reaction, definition of the spliceosome and identification of its components, and biochemical analysis of the various splicing complexes and their regulation. Structural information is central to mechanistic understanding of pre-mRNA splicing by the spliceosome. X-ray crystallography of the spliceosomal components and subcomplexes is complemented by electron microscopy of the intact spliceosome. In this Review, I discuss recent atomic-resolution structures of the intact spliceosome at different stages of the splicing cycle. These structures have provided considerable mechanistic insight into pre-mRNA splicing and have corroborated and explained a large body of genetic and biochemical data. Together, the structural data have proved that the spliceosome is a protein-directed metalloribozyme.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Berget, S. M., Moore, C. & Sharp, P. A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl Acad. Sci. USA 74, 3171–3175 (1977).
Chow, L. T., Gelinas, R. E., Broker, T. R. & Roberts, R. J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12, 1–8 (1977).
Lerner, M. R. & Steitz, J. A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc. Natl Acad. Sci. USA 76, 5495–5499 (1979).
Lerner, M. R., Boyle, J. A., Mount, S. M., Wolin, S. L. & Steitz, J. A. Are snRNPs involved in splicing? Nature 283, 220–224 (1980).
Rogers, J. & Wall, R. A mechanism for RNA splicing. Proc. Natl Acad. Sci. USA 77, 1877–1879 (1980).
Hinterberger, M., Pettersson, I. & Steitz, J. A. Isolation of small nuclear ribonucleoproteins containing U1, U2, U4, U5, and U6 RNAs. J. Biol. Chem. 258, 2604–2613 (1983).
Mount, S. M., Pettersson, I., Hinterberger, M., Karmas, A. & Steitz, J. A. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell 33, 509–518 (1983).
Padgett, R. A., Mount, S. M., Steitz, J. A. & Sharp, P. A. Splicing of messenger RNA precursors is inhibited by antisera to small nuclear ribonucleoprotein. Cell 35, 101–107 (1983).
Yang, V. W., Lerner, M. R., Steitz, J. A. & Flint, S. J. A small nuclear ribonucleoprotein is required for splicing of adenoviral early RNA sequences. Proc. Natl Acad. Sci. USA 78, 1371–1375 (1981).
DiMaria, P. R., Kaltwasser, G. & Goldenberg, C. J. Partial purification and properties of a pre-mRNA splicing activity. J. Biol. Chem. 260, 1096–1102 (1985).
Kramer, A., Keller, W., Appel, B. & Luhrmann, R. The 5′ terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell 38, 299–307 (1984).
Black, D. L., Chabot, B. & Steitz, J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell 42, 737–750 (1985).
Krainer, A. R. & Maniatis, T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell 42, 725–736 (1985).
Berget, S. M. & Robberson, B. L. U1, U2, and U4/U6 small nuclear ribonucleoproteins are required for in vitro splicing but not polyadenylation. Cell 46, 691–696 (1986).
Grabowski, P. J. & Sharp, P. A. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science 233, 1294–1299 (1986).
Pikielny, C. W. & Rosbash, M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell 45, 869–877 (1986).
Goldenberg, C. J. & Hauser, S. D. Accurate and efficient in vitro splicing of purified precursor RNAs specified by early region 2 of the adenovirus 2 genome. Nucleic Acids Res. 11, 1337–1348 (1983).
Hernandez, N. & Keller, W. Splicing of in vitro synthesized messenger RNA precursors in HeLa cell extracts. Cell 35, 89–99 (1983).
Kole, R. & Weissman, S. M. Accurate in vitro splicing of human beta-globin RNA. Nucleic Acids Res. 10, 5429–5445 (1982).
Padgett, R. A., Hardy, S. F. & Sharp, P. A. Splicing of adenovirus RNA in a cell-free transcription system. Proc. Natl Acad. Sci. USA 80, 5230–5234 (1983).
Hardy, S. F., Grabowski, P. J., Padgett, R. A. & Sharp, P. A. Cofactor requirements of splicing of purified messenger RNA precursors. Nature 308, 375–377 (1984).
Krainer, A. R., Maniatis, T., Ruskin, B. & Green, M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36, 993–1005 (1984).
Grabowski, P. J., Padgett, R. A. & Sharp, P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell 37, 415–427 (1984).
Padgett, R. A., Konarska, M. M., Grabowski, P. J., Hardy, S. F. & Sharp, P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science 225, 898–903 (1984).
Ruskin, B., Krainer, A. R., Maniatis, T. & Green, M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38, 317–331 (1984).
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993).
Sontheimer, E. J., Sun, S. & Piccirilli, J. A. Metal ion catalysis during splicing of premessenger RNA. Nature 388, 801–805 (1997).
Yean, S. L., Wuenschell, G., Termini, J. & Lin, R. J. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408, 881–884 (2000).
Fica, S. M. et al. RNA catalyses nuclear pre-mRNA splicing. Nature 503, 229–234 (2013).
Toor, N., Keating, K. S., Taylor, S. D. & Pyle, A. M. Crystal structure of a self-spliced group II intron. Science 320, 77–82 (2008).
Keating, K. S., Toor, N., Perlman, P. S. & Pyle, A. M. A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16, 1–9 (2010).
Brody, E. & Abelson, J. The “spliceosome”: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 228, 963–967 (1985).
Grabowski, P. J., Seiler, S. R. & Sharp, P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell 42, 345–353 (1985).
Frendewey, D. & Keller, W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell 42, 355–367 (1985).
Aebi, M., Hornig, H., Padgett, R. A., Reiser, J. & Weissmann, C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47, 555–565 (1986).
Vijayraghavan, U. et al. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 5, 1683–1695 (1986).
Newman, A. J., Lin, R. J., Cheng, S. C. & Abelson, J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell 42, 335–344 (1985).
Lamond, A. I., Konarska, M. M. & Sharp, P. A. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1, 532–543 (1987).
Konarska, M. M. & Sharp, P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46, 845–855 (1986).
Pikielny, C. W., Rymond, B. C. & Rosbash, M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature 324, 341–345 (1986).
Konarska, M. M. & Sharp, P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell 49, 763–774 (1987).
Cheng, S. C. & Abelson, J. Spliceosome assembly in yeast. Genes Dev. 1, 1014–1027 (1987).
Bindereif, A. & Green, M. R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 6, 2415–2424 (1987).
Wahl, M. C., Will, C. L. & Luhrmann, R. The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 (2009).
Guthrie, C. & Patterson, B. Spliceosomal snRNAs. Annu. Rev. Genet. 22, 387–419 (1988).
Bringmann, P. & Luhrmann, R. Purification of the individual snRNPs U1, U2, U5 and U4/U6 from HeLa cells and characterization of their protein constituents. EMBO J. 5, 3509–3516 (1986).
Lossky, M., Anderson, G. J., Jackson, S. P. & Beggs, J. Identification of a yeast snRNP protein and detection of snRNP-snRNP interactions. Cell 51, 1019–1026 (1987).
Jackson, S. P., Lossky, M. & Beggs, J. D. Cloning of the RNA8 gene of Saccharomyces cerevisiae, detection of the RNA8 protein, and demonstration that it is essential for nuclear pre-mRNA splicing. Mol. Cell. Biol. 8, 1067–1075 (1988).
Tarn, W. Y. et al. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13, 2421–2431 (1994).
Chan, S. P., Kao, D. I., Tsai, W. Y. & Cheng, S. C. The Prp19p-associated complex in spliceosome activation. Science 302, 279–282 (2003).
Chabot, B. & Steitz, J. A. Multiple interactions between the splicing substrate and small nuclear ribonucleoproteins in spliceosomes. Mol. Cell. Biol. 7, 281–293 (1987).
Parker, R., Siliciano, P. G. & Guthrie, C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49, 229–239 (1987).
Newman, A. & Norman, C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell 65, 115–123 (1991).
Newman, A. J. & Norman, C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 68, 743–754 (1992).
Madhani, H. D. & Guthrie, C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71, 803–817 (1992).
Wassarman, D. A. & Steitz, J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science 257, 1918–1925 (1992).
Wyatt, J. R., Sontheimer, E. J. & Steitz, J. A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 6, 2542–2553 (1992).
Lesser, C. F. & Guthrie, C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science 262, 1982–1988 (1993).
Sontheimer, E. J. & Steitz, J. A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262, 1989–1996 (1993).
Kandels-Lewis, S. & Seraphin, B. Involvement of U6 snRNA in 5′ splice site selection. Science 262, 2035–2039 (1993).
Newman, A. J., Teigelkamp, S. & Beggs, J. D. snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1, 968–980 (1995).
Anokhina, M. et al. RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. EMBO J. 32, 2804–2818 (2013).
Jankowsky, E. RNA helicases at work: binding and rearranging. Trends Biochem. Sci. 36, 19–29 (2011).
Cordin, O., Hahn, D. & Beggs, J. D. Structure, function and regulation of spliceosomal RNA helicases. Curr. Opin. Cell Biol. 24, 431–438 (2012).
Staley, J. P. & Guthrie, C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315–326 (1998).
Raghunathan, P. L. & Guthrie, C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8, 847–855 (1998).
Laggerbauer, B., Achsel, T. & Luhrmann, R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl Acad. Sci. USA 95, 4188–4192 (1998).
Chen, J. H. & Lin, R. J. The yeast PRP2 protein, a putative RNA-dependent ATPase, shares extensive sequence homology with two other pre-mRNA splicing factors. Nucleic Acids Res. 18, 6447 (1990).
King, D. S. & Beggs, J. D. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 18, 6559–6564 (1990).
Kim, S. H. & Lin, R. J. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 16, 6810–6819 (1996).
Kim, S. H., Smith, J., Claude, A. & Lin, R. J. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 11, 2319–2326 (1992).
Burgess, S., Couto, J. R. & Guthrie, C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell 60, 705–717 (1990).
Schwer, B. & Guthrie, C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature 349, 494–499 (1991).
Company, M., Arenas, J. & Abelson, J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349, 487–493 (1991).
Semlow, D. R., Blanco, M. R., Walter, N. G. & Staley, J. P. Spliceosomal DEAH-box ATPases remodel pre-mRNA to activate alternative splice sites. Cell 164, 985–998 (2016).
Semlow, D. R. & Staley, J. P. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem. Sci. 37, 263–273 (2012).
Weber, G., Trowitzsch, S., Kastner, B., Luhrmann, R. & Wahl, M. C. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 29, 4172–4184 (2010).
Pomeranz Krummel, D. A., Oubridge, C., Leung, A. K., Li, J. & Nagai, K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature 458, 475–480 (2009).
Kondo, Y., Oubridge, C., van Roon, A. M. & Nagai, K. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife http://dx.doi.org/10.7554/eLife.04986 (2015).
Price, S. R., Evans, P. R. & Nagai, K. Crystal structure of the spliceosomal U2B”-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature 394, 645–650 (1998).
Sickmier, E. A. et al. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell 23, 49–59 (2006).
Lin, P. C. & Xu, R. M. Structure and assembly of the SF3a splicing factor complex of U2 snRNP. EMBO J. 31, 1579–1590 (2012).
Jenkins, J. L., Agrawal, A. A., Gupta, A., Green, M. R. & Kielkopf, C. L. U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs. Nucleic Acids Res. 41, 3859–3873 (2013).
Yoshida, H. et al. A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev. 29, 1649–1660 (2015).
Leung, A. K., Nagai, K. & Li, J. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473, 536–539 (2011).
Zhou, L. et al. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature 506, 116–120 (2014).
Montemayor, E. J. et al. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-A resolution. Nat. Struct. Mol. Biol. 21, 544–551 (2014).
Galej, W. P., Oubridge, C., Newman, A. J. & Nagai, K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 493, 638–643 (2013).
Mozaffari-Jovin, S. et al. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science 341, 80–84 (2013).
Nguyen, T. H. et al. Structural basis of Brr2–Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure 21, 910–919 (2013).
Cretu, C. et al. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol. Cell 64, 307–319 (2016).
Zhou, Z., Sim, J., Griffith, J. & Reed, R. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl Acad. Sci. USA 99, 12203–12207 (2002).
Jurica, M. S., Licklider, L. J., Gygi, S. R., Grigorieff, N. & Moore, M. J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8, 426–439 (2002).
Luhrmann, R. & Stark, H. Structural mapping of spliceosomes by electron microscopy. Curr. Opin. Struct. Biol. 19, 96–102 (2009).
Behzadnia, N. et al. Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26, 1737–1748 (2007).
Furman, E. & Glitz, D. G. Purification of the spliceosome A-complex and its visualization by electron microscopy. J. Biol. Chem. 270, 15515–15522 (1995).
Boehringer, D. et al. Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat. Struct. Mol. Biol. 11, 463–468 (2004).
Wolf, E. et al. Exon, intron and splice site locations in the spliceosomal B complex. EMBO J. 28, 2283–2292 (2009).
Deckert, J. et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26, 5528–5543 (2006).
Bessonov, S. et al. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16, 2384–2403 (2010).
Golas, M. M. et al. 3D cryo-EM structure of an active step I spliceosome and localization of its catalytic core. Mol. Cell 40, 927–938 (2010).
Jurica, M. S., Sousa, D., Moore, M. J. & Grigorieff, N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat. Struct. Mol. Biol. 11, 265–269 (2004).
Ilagan, J. O., Chalkley, R. J., Burlingame, A. L. & Jurica, M. S. Rearrangements within human spliceosomes captured after exon ligation. RNA 19, 400–412 (2013).
Fabrizio, P. et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell 36, 593–608 (2009).
Ohi, M. D., Ren, L., Wall, J. S., Gould, K. L. & Walz, T. Structural characterization of the fission yeast U5. U2/U6 spliceosome complex. Proc. Natl Acad. Sci. USA 104, 3195–3200 (2007).
Chen, W. et al. Endogenous U2. U5. U6 snRNA complexes in S. pombe are intron lariat spliceosomes. RNA 20, 308–320 (2014).
Nguyen, T. H. et al. The architecture of the spliceosomal U4/U6. U5 tri-snRNP. Nature 523, 47–52 (2015).
Yan, C. et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349, 1182–1191 (2015).
Hang, J., Wan, R., Yan, C. & Shi, Y. Structural basis of pre-mRNA splicing. Science 349, 1191–1198 (2015).
Wan, R. et al. The 3.8 A structure of the U4/U6. U5 tri-snRNP: insights into spliceosome assembly and catalysis. Science 351, 466–475 (2016).
Nguyen, T. H. et al. Cryo-EM structure of the yeast U4/U6. U5 tri-snRNP at 3.7 A resolution. Nature 530, 298–302 (2016).
Agafonov, D. E. et al. Molecular architecture of the human U4/U6. U5 tri-snRNP. Science 351, 1416–1420 (2016).
Yan, C., Wan, R., Bai, R., Huang, G. & Shi, Y. Structure of a yeast activated spliceosome at 3.5 A resolution. Science 353, 904–911 (2016).
Wan, R., Yan, C., Bai, R., Huang, G. & Shi, Y. Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science 353, 895–904 (2016).
Galej, W. P. et al. Cryo-EM structure of the spliceosome immediately after branching. Nature 537, 197–201 (2016).
Yan, C., Wan, R., Bai, R., Huang, G. & Shi, Y. Structure of a yeast step II catalytically activated spliceosome. Science 355, 149–155 (2017).
Fica, S. M. et al. Structure of a spliceosome remodelled for exon ligation. Nature 542, 377–380 (2017).
Rauhut, R. et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353, 1399–1405 (2016).
Bertram, K. et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542, 318–323 (2017).
Fica, S. M., Mefford, M. A., Piccirilli, J. A. & Staley, J. P. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat. Struct. Mol. Biol. 21, 464–471 (2014).
Robart, A. R., Chan, R. T., Peters, J. K., Rajashankar, K. R. & Toor, N. Crystal structure of a eukaryotic group II intron lariat. Nature 514, 193–197 (2014).
Grainger, R. J. & Beggs, J. D. Prp8 protein: at the heart of the spliceosome. RNA 11, 533–557 (2005).
Turner, I. A., Norman, C. M., Churcher, M. J. & Newman, A. J. Dissection of Prp8 protein defines multiple interactions with crucial RNA sequences in the catalytic core of the spliceosome. RNA 12, 375–386 (2006).
Galej, W. P., Nguyen, T. H., Newman, A. J. & Nagai, K. Structural studies of the spliceosome: zooming into the heart of the machine. Curr. Opin. Struct. Biol. 25, 57–66 (2014).
Yang, K., Zhang, L., Xu, T., Heroux, A. & Zhao, R. Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc. Natl Acad. Sci. USA 105, 13817–13822 (2008).
Garrey, S. M. et al. A homolog of lariat-debranching enzyme modulates turnover of branched RNA. RNA 20, 1337–1348 (2014).
Rasche, N. et al. Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre. EMBO J. 31, 1591–1604 (2012).
Hahn, C. N. & Scott, H. S. Spliceosome mutations in hematopoietic malignancies. Nat. Genet. 44, 9–10 (2012).
Darman, R. B. et al. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 13, 1033–1045 (2015).
Alsafadi, S. et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 7, 10615 (2016).
Jacquier, A. & Michel, F. Base-pairing interactions involving the 5′ and 3′-terminal nucleotides of group II self-splicing introns. J. Mol. Biol. 213, 437–447 (1990).
Chanfreau, G. & Jacquier, A. Catalytic site components common to both splicing steps of a group II intron. Science 266, 1383–1387 (1994).
Umen, J. G. & Guthrie, C. The second catalytic step of pre-mRNA splicing. RNA 1, 869–885 (1995).
Luukkonen, B. G. & Seraphin, B. The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J. 16, 779–792 (1997).
Collins, C. A. & Guthrie, C. Genetic interactions between the 5′ and 3′ splice site consensus sequences and U6 snRNA during the second catalytic step of pre-mRNA splicing. RNA 7, 1845–1854 (2001).
Frank, D. & Guthrie, C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 6, 2112–2124 (1992).
Umen, J. G. & Guthrie, C. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA 1, 584–597 (1995).
Umen, J. G. & Guthrie, C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics 143, 723–739 (1996).
Chua, K. & Reed, R. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature 402, 207–210 (1999).
Chen, W. & Moore, M. J. The spliceosome: disorder and dynamics defined. Curr. Opin. Struct. Biol. 24, 141–149 (2014).
Ohi, M. D. & Gould, K. L. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8, 798–815 (2002).
Ohrt, T. et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA 18, 1244–1256 (2012).
Grainger, R. J., Barrass, J. D., Jacquier, A., Rain, J. C. & Beggs, J. D. Physical and genetic interactions of yeast Cwc21p, an ortholog of human SRm300/SRRM2, suggest a role at the catalytic center of the spliceosome. RNA 15, 2161–2173 (2009).
Warkocki, Z. et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 16, 1237–1243 (2009).
Chiu, Y. F. et al. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol. Cell. Biol. 29, 5671–5678 (2009).
Krishnan, R. et al. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nat. Struct. Mol. Biol. 20, 1450–1457 (2013).
Ohrt, T. et al. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA 19, 902–915 (2013).
James, S. A., Turner, W. & Schwer, B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8, 1068–1077 (2002).
Zhang, X. & Schwer, B. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 25, 2146–2152 (1997).
Santos, K. F. et al. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc. Natl Acad. Sci. USA 109, 17418–17423 (2012).
Hahn, D., Kudla, G., Tollervey, D. & Beggs, J. D. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 26, 2408–2421 (2012).
Huang, Y. H., Chung, C. S., Kao, D. I., Kao, T. C. & Cheng, S. C. Sad1 counteracts Brr2-mediated dissociation of U4/U6. U5 in tri-snRNP homeostasis. Mol. Cell. Biol. 34, 210–220 (2014).
McPheeters, D. S. & Muhlenkamp, P. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 23, 4174–4186 (2003).
Schneider, C. et al. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3′ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLoS Genet. 11, e1005539 (2015).
Edwalds-Gilbert, G., Kim, D. H., Silverman, E. & Lin, R. J. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA 10, 210–220 (2004).
Liu, H. L. & Cheng, S. C. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol. Cell. Biol. 32, 5056–5066 (2012).
Warkocki, Z. et al. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 29, 94–107 (2015).
Teigelkamp, S., McGarvey, M., Plumpton, M. & Beggs, J. D. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 13, 888–897 (1994).
Lardelli, R. M., Thompson, J. X., Yates, J. R. 3rd & Stevens, S. W. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16, 516–528 (2010).
Schwer, B. & Guthrie, C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 11, 5033–5039 (1992).
Tseng, C. K., Liu, H. L. & Cheng, S. C. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17, 145–154 (2011).
Schwer, B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol. Cell 30, 743–754 (2008).
Schwer, B. & Gross, C. H. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17, 2086–2094 (1998).
Sharp, P. A. On the origin of RNA splicing and introns. Cell 42, 397–400 (1985).
Cech, T. R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell 44, 207–210 (1986).
Maroney, P. A., Romfo, C. M. & Nilsen, T. W. Functional recognition of 5′ splice site by U4/U6. U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol. Cell 6, 317–328 (2000).
Tsai, R. T. et al. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19, 2991–3003 (2005).
Fourmann, J. B., Tauchert, M. J., Ficner, R., Fabrizio, P. & Luhrmann, R. Regulation of Prp43-mediated disassembly of spliceosomes by its cofactors Ntr1 and Ntr2. Nucleic Acids Res. 45, 4068–4080 (2017).
Acknowledgements
The author apologizes to those colleagues whose important work is not cited in this review owing to space limitation or inadvertent omission. The author thanks R. Wan, R. Bai, J. Hang, X. Zhang, X. Zhan and C. Yan for preparation of the figures and suggestions on text and references. This work was supported by funds from the Beijing Advanced Innovation Center for Structural Biology at Tsinghua University, Beijing, China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Cryo-EM studies of the spliceosome (PDF 117 kb)
Supplementary information S2 (figure)
Recognition of the exon sequences. (PDF 8180 kb)
Supplementary information S3 (figure)
Distinct classes of spliceosomal proteins. (PDF 252 kb)
Supplementary information S4 (figure)
Comparison of the catalytic centers between the spliceosome and the self-splicing group IIB intron. (PDF 3340 kb)
Glossary
- Intron lariat
-
A lasso-like intron structure formed by the first transesterification of the splicing cycle.
- Branch point sequence
-
(BPS). Also known as a branch site. A conserved intron sequence, which contains an invariant adenosine nucleotide that serves as the nucleophile for the branching reaction.
- SN2-type transesterification
-
Transesterification in which one ester bond is broken and another ester bond is formed synchronously; named for substitution nucleophilic and bi-molecular transesterification.
- Group II self-splicing introns
-
A large class of self-splicing catalytic RNAs found in bacteria, fungi and plants.
- MS2-tagged
-
RNA prepared for affinity purification using a technique based on the interactions of the MS2 bacteriophage coat protein with a stem–loop RNA structure from the phage genome.
- Lariat junction
-
The three-way junction in which a 2′ to 5′ linkage is formed between the ribose 2′-OH of an adenosine nucleotide in the intron branch site and the phosphate of a guanine nucleotide at the 5′ end of the intron 5′ splice site.
- HEAT repeats
-
Repetitive motifs frequently found in signalling proteins; each HEAT repeat comprises a pair of anti-parallel α-helices, linked by a short loop.
- HAT repeats
-
Repetitive motifs implicated in RNA processing; each HAT repeat comprises two α-helices.
- WD40 domain
-
Also known as β-propeller or WD repeats. A type of circular solenoid protein domain that consists of 5–8 short structural motifs of approximately 40 amino acids each, often terminating in a tryptophan–aspartic acid (WD) dipeptide.
- Sm ring
-
Also known as heptameric Sm complex. A doughnut-shaped structure formed by seven distinct Sm proteins, with single-stranded RNA bound within the hollow centre.
Rights and permissions
About this article
Cite this article
Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol 18, 655–670 (2017). https://doi.org/10.1038/nrm.2017.86
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2017.86
This article is cited by
-
A high heterozygosity genome assembly of Aedes albopictus enables the discovery of the association of PGANT3 with blood-feeding behavior
BMC Genomics (2024)
-
The spliceosome-associated protein CWC15 promotes miRNA biogenesis in Arabidopsis
Nature Communications (2024)
-
Importance of pre-mRNA splicing and its study tools in plants
Advanced Biotechnology (2024)
-
Regulation of adult stem cell quiescence and its functions in the maintenance of tissue integrity
Nature Reviews Molecular Cell Biology (2023)
-
A guide to membraneless organelles and their various roles in gene regulation
Nature Reviews Molecular Cell Biology (2023)