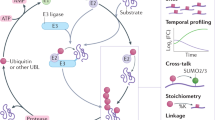
Key Points
-
Small ubiquitin-like modifier (SUMO)ylation is a ubiquitin-like post-translational modification that is technically challenging to study by mass spectrometry-based proteomics, owing to low modification stoichiometry and incompatibility with standard mass spectrometry approaches.
-
Over the past decade, several methods were developed to allow the identification of sumoylated proteins. Recently, novel methods were established that enable the identification of site-specific sumoylation, leading to the identification of thousands of modified Lys residues.
-
We have compiled and evaluated data of sumoylated proteins and sites from 22 proteomic studies and generated the most comprehensive sumoylation database to date.
-
The most abundantly sumoylated proteins are some of the most functionally interconnected proteins, indicating that sumoylation preferentially targets specific pathways and protein complexes and acts as a group modifier.
-
The regulation of the SUMO signal is more complex than previously appreciated, owing to the post-translational modification of SUMO family members by phosphorylation, acetylation and ubiquitylation.
-
With the availability of increasingly potent proteomic methods and equipment, our understanding of sumoylation and its functions can now be developed to match that of ubiquitylation, acetylation and phosphorylation.
Abstract
Small ubiquitin-like modifiers (SUMOs) are essential for the regulation of several cellular processes and are potential therapeutic targets owing to their involvement in diseases such as cancer and Alzheimer disease. In the past decade, we have witnessed a rapid expansion of proteomic approaches for identifying sumoylated proteins, with recent advances in detecting site-specific sumoylation. In this Analysis, we combined all human SUMO proteomics data currently available into one cohesive database. We provide proteomic evidence for sumoylation of 3,617 proteins at 7,327 sumoylation sites, and insight into SUMO group modification by clustering the sumoylated proteins into functional networks. The data support sumoylation being a frequent protein modification (on par with other major protein modifications) with multiple nuclear functions, including in transcription, mRNA processing, DNA replication and the DNA-damage response.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Golebiowski, F. et al. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2, ra24 (2009).
Hendriks, I. A. et al. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 21, 927–936 (2014). The first site-specific proteomics paper reporting more than 1,000 sumoylation sites in standard growth conditions, and more than 4,000 sumoylation sites in total.
Schimmel, J. et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell 53, 1053–1066 (2014).
Miura, K., Jin, J. B. & Hasegawa, P. M. Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10, 495–502 (2007).
Geiss-Friedlander, R. & Melchior, F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 (2007).
Flotho, A. & Melchior, F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 (2013).
Ulrich, H. D. & Walden, H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 11, 479–489 (2010).
Hickey, C. M., Wilson, N. R. & Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766 (2012).
Jackson, S. P. & Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 (2013).
Eifler, K. & Vertegaal, A. C. Mapping the SUMOylated landscape. FEBS J. 282, 3669–3680 (2015).
Eifler, K. & Vertegaal, A. C. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem. Sci. 40, 779–793 (2015).
Kessler, J. D. et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335, 348–353 (2012). The involvement of SUMO in diseases such as cancer is exemplified by the ability of SUMO to orchestrate Myc-driven tumorigenesis.
Lee, Y. J. & Hallenbeck, J. M. SUMO and ischemic tolerance. Neuromolecular Med. 15, 771–781 (2013).
Wang, Y. & Dasso, M. SUMOylation and deSUMOylation at a glance. J. Cell Sci. 122, 4249–4252 (2009).
Guo, D. et al. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem. Biophys. Res. Commun. 337, 1308–1318 (2005).
Becker, J. et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20, 525–531 (2013). The first proteomics screen to identify hundreds of endogenous SUMO target proteins using immunoprecipitation.
Matic, I. et al. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics 7, 132–144 (2008).
Tatham, M. H. et al. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276, 35368–35374 (2001).
Seeler, J. S. & Dejean, A. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4, 690–699 (2003).
Kamitani, T., Nguyen, H. P. & Yeh, E. T. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J. Biol. Chem. 272, 14001–14004 (1997).
Shiio, Y. & Eisenman, R. N. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. USA 100, 13225–13230 (2003).
Stielow, B. et al. Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol. Cell 29, 742–754 (2008).
Uchimura, Y. et al. Involvement of SUMO modification in. J. Biol. Chem. 281, 23180–23190 (2006).
Ishov, A. M. et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147, 221–234 (1999).
Zhong, S. et al. Role of SUMO-1-modified PML in nuclear body formation. Blood 95, 2748–2752 (2000).
Muller, S. & Dejean, A. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73, 5137–5143 (1999).
Shen, T. H., Lin, H. K., Scaglioni, P. P., Yung, T. M. & Pandolfi, P. P. The mechanisms of PML-nuclear body formation. Mol. Cell 24, 331–339 (2006).
Bernardi, R. & Pandolfi, P. P. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 22, 9048–9057 (2003).
Dellaire, G. & Bazett-Jones, D. P. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26, 963–977 (2004).
Bergink, S. & Jentsch, S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 (2009).
Hardeland, U., Steinacher, R., Jiricny, J. & Schar, P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21, 1456–1464 (2002).
Stelter, P. & Ulrich, H. D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 (2003).
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002).
Morris, J. R. et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 (2009).
Galanty, Y. et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 (2009).
Yin, Y. et al. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 (2012).
Galanty, Y., Belotserkovskaya, R., Coates, J. & Jackson, S. P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, 1179–1195 (2012).
Vyas, R. et al. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ. 20, 490–502 (2013).
Ouyang, K. J. et al. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 7, e1000252 (2009).
Pichler, A. et al. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat. Struct. Mol. Biol. 12, 264–269 (2005).
Steinacher, R. & Schar, P. Functionality of human thymine DNA glycosylase requires SUMO-regulated changes in protein conformation. Curr. Biol. 15, 616–623 (2005).
Hecker, C. M., Rabiller, M., Haglund, K., Bayer, P. & Dikic, I. Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 (2006).
Sun, H. & Hunter, T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J. Biol. Chem. 287, 42071–42083 (2012).
Song, J., Durrin, L. K., Wilkinson, T. A., Krontiris, T. G. & Chen, Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl Acad. Sci. USA 101, 14373–14378 (2004).
Keusekotten, K. et al. Multivalent interactions of the SUMO-interaction motifs in RING finger protein 4 determine the specificity for chains of the SUMO. Biochem. J. 457, 207–214 (2014).
Merrill, J. C. et al. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS ONE 5, e8794 (2010).
Prudden, J. et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 (2007).
Perry, J. J., Tainer, J. A. & Boddy, M. N. A. SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 33, 201–208 (2008).
Wang, Z. & Prelich, G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell. Biol. 29, 1694–1706 (2009).
Nagai, S., Davoodi, N. & Gasser, S. M. Nuclear organization in genome stability: SUMO connections. Cell Res. 21, 474–485 (2011).
Sun, H., Leverson, J. D. & Hunter, T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 (2007).
Tatham, M. H. et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 (2008).
Weisshaar, S. R. et al. Arsenic trioxide stimulates SUMO-2/3 modification leading to RNF4-dependent proteolytic targeting of PML. FEBS Lett. 582, 3174–3178 (2008).
Geoffroy, M. C., Jaffray, E. G., Walker, K. J. & Hay, R. T. Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol. Biol. Cell 21, 4227–4239 (2010).
Lallemand-Breitenbach, V. et al. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10, 547–555 (2008).
Kosoy, A., Calonge, T. M., Outwin, E. A. & O'Connell, M. J. Fission yeast Rnf4 homologs are required for DNA repair. J. Biol. Chem. 282, 20388–20394 (2007).
Guzzo, C. M. et al. RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci. Signal. 5, ra88 (2012).
Silver, H. R., Nissley, J. A., Reed, S. H., Hou, Y. M. & Johnson, E. S. A role for SUMO in nucleotide excision repair. DNA Repair (Amst.) 10, 1243–1251 (2011).
Psakhye, I. & Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820 (2012). An excellent example of SUMO group modification during the DDR in yeast.
Raman, N., Nayak, A. & Muller, S. The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma 122, 475–485 (2013).
Sahin, U. et al. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 204, 931–945 (2014).
Gonzalez-Prieto, R., Cuijpers, S. A., Luijsterburg, M. S., van Attikum, H. & Vertegaal, A. C. SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. EMBO Rep. 16, 512–519 (2015).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Pandey, A. & Mann, M. Proteomics to study genes and genomes. Nature 405, 837–846 (2000).
Mann, M. & Jensen, O. N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 (2003).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Olsen, J. V. & Mann, M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 12, 3444–3452 (2013).
Choudhary, C. & Mann, M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 11, 427–439 (2010).
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009).
Huttlin, E. L. et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010).
Olsen, J. V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006).
Guo, A. et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–387 (2013).
Emanuele, M. J. et al. Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 (2011).
Wagner, S. A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284 (2011).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Tammsalu, T. et al. Proteome-wide identification of SUMO2 modification sites. Sci. Signal. 7, rs2 (2014). The first site-specific SUMO proteomics study to report more than 1,000 sumoylation sites.
Xiao, Z. et al. System-wide analysis of SUMOylation dynamics in response to replication stress reveals novel SUMO target proteins and acceptor lysines relevant for genome stability. Mol. Cell. Proteomics 14, 1419–1434 (2015).
Hendriks, I. A., Treffers, L. W., Verlaan-de Vries, M., Olsen, J. V. & Vertegaal, A. C. SUMO-2 orchestrates chromatin modifiers in response to DNA damage. Cell Rep. 10, 1778–1791 (2015).
Impens, F., Radoshevich, L., Cossart, P. & Ribet, D. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl Acad. Sci. USA 111, 12432–12437 (2014).
Lamoliatte, F. et al. Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat. Commun. 5, 5409 (2014).
Hendriks, I. A., D'Souza, R. C., Chang, J. G., Mann, M. & Vertegaal, A. C. System-wide identification of wild-type SUMO-2 conjugation sites. Nat. Commun. 6, 7289 (2015). Describes the development of PRISM, the first proteomic method to be able to identify hundreds of sumoylation sites without relying on the use of mutant SUMO.
Mahajan, R., Gerace, L. & Melchior, F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140, 259–270 (1998).
Eisenhardt, N. et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat. Struct. Mol. Biol. 22, 959–967 (2015). Identifies ZNF451, a newly discovered enzyme with SUMO E3 ligase activity that is able to elongate SUMO chains and mediate sumoylation dynamics in response to cellular stress.
Cappadocia, L., Pichler, A. & Lima, C. D. Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase. Nat. Struct. Mol. Biol. 22, 968–975 (2015).
Schimmel, J. et al. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell. Proteomics 7, 2107–2122 (2008).
Pinto, M. P. et al. Heat shock induces a massive but differential inactivation of SUMO-specific proteases. Biochim. Biophys. Acta 1823, 1958–1966 (2012).
Guo, L. et al. A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol. Cell 55, 15–30 (2014).
Bursomanno, S. et al. Proteome-wide analysis of SUMO2 targets in response to pathological DNA replication stress in human cells. DNA Repair (Amst.) 25, 84–96 (2015).
Seifert, A., Schofield, P., Barton, G. J. & Hay, R. T. Proteotoxic stress reprograms the chromatin landscape of SUMO modification. Sci. Signal. 8, rs7 (2015).
Li, T. et al. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl Acad. Sci. USA 101, 8551–8556 (2004).
Vassileva, M. T. & Matunis, M. J. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell. Biol. 24, 3623–3632 (2004).
Pelisch, F., Pozzi, B., Risso, G., Munoz, M. J. & Srebrow, A. DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation. J. Biol. Chem. 287, 30789–30799 (2012).
Lee, S. W. et al. SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J. 31, 4441–4452 (2012).
Panse, V. G. et al. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 7, 1311–1321 (2006).
Finkbeiner, E., Haindl, M. & Muller, S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 30, 1067–1078 (2011).
Westman, B. J. et al. A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol. Cell 39, 618–631 (2010).
Zemp, I. & Kutay, U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581, 2783–2793 (2007).
Hang, J. & Dasso, M. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277, 19961–19966 (2002).
Lewis, A., Felberbaum, R. & Hochstrasser, M. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J. Cell Biol. 178, 813–827 (2007).
David, G., Neptune, M. A. & DePinho, R. A. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277, 23658–23663 (2002).
Yang, S. H. & Sharrocks, A. D. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13, 611–617 (2004).
Kirsh, O. et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21, 2682–2691 (2002).
Bueno, M. T. & Richard, S. SUMOylation negatively modulates target gene occupancy of the KDM5B, a histone lysine demethylase. Epigenetics 8, 1162–1175 (2013).
Sriramachandran, A. M. & Dohmen, R. J. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 1843, 75–85 (2014).
Ryu, H., Gygi, S. P., Azuma, Y., Arnaoutov, A. & Dasso, M. SUMOylation of Psmd1 controls Adrm1 interaction with the proteasome. Cell Rep. 7, 1842–1848 (2014).
Nacerddine, K. et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9, 769–779 (2005).
Myatt, S. S. et al. SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene 33, 4316–4329 (2013).
Zhang, X. D. et al. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 29, 729–741 (2008).
Wang, Q. E. et al. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 33, 4023–4034 (2005).
Luo, K., Zhang, H., Wang, L., Yuan, J. & Lou, Z. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 31, 3008–3019 (2012).
Gonzalez-Prieto, R., Cuijpers, S. A., Kumar, R., Hendriks, I. A. & Vertegaal, A. C. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 14, 1859–1872 (2015).
Sabo, A., Doni, M. & Amati, B. SUMOylation of Myc-family proteins. PLoS ONE 9, e91072 (2014).
Schou, J., Kelstrup, C. D., Hayward, D. G., Olsen, J. V. & Nilsson, J. Comprehensive identification of SUMO2/3 targets and their dynamics during mitosis. PLoS ONE 9, e100692 (2014).
Cubenas-Potts, C. et al. Identification of SUMO-2/3-modified proteins associated with mitotic chromosomes. Proteomics 15, 763–772 (2015).
Bursomanno, S., McGouran, J. F., Kessler, B. M., Hickson, I. D. & Liu, Y. Regulation of SUMO2 target proteins by the proteasome in human cells exposed to replication stress. J. Proteome Res. 14, 1687–1699 (2015).
Rodriguez, M. S., Dargemont, C. & Hay, R. T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659 (2001).
Bernier-Villamor, V., Sampson, D. A., Matunis, M. J. & Lima, C. D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345–356 (2002).
Matic, I. et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652 (2010).
Yang, S. H., Galanis, A., Witty, J. & Sharrocks, A. D. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 25, 5083–5093 (2006).
Hietakangas, V. et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl Acad. Sci. USA 103, 45–50 (2006).
Kubota, Y., O'Grady, P., Saito, H. & Takekawa, M. Oncogenic Ras abrogates MEK SUMOylation that suppresses the ERK pathway and cell transformation. Nat. Cell Biol. 13, 282–291 (2011).
Girdwood, D. et al. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11, 1043–1054 (2003).
Ullmann, R., Chien, C. D., Avantaggiati, M. L. & Muller, S. An acetylation switch regulates SUMO-dependent protein interaction networks. Mol. Cell 46, 759–770 (2012).
Wagner, S. A. et al. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics 11, 1578–1585 (2012).
Danielsen, J. M. et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10, M110.003590 (2011).
Olsen, J. V. et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 (2010).
Choudhary, C. et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol. Cell 36, 326–339 (2009).
Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res. 26, 399–422 (2016).
Herhaus, L. & Dikic, I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 16, 1071–1083 (2015).
Ordureau, A. et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl Acad. Sci. USA 112, 6637–6642 (2015).
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Vertegaal, A. C. SUMO chains: polymeric signals. Biochem. Soc. Trans. 38, 46–49 (2010).
Desterro, J. M., Rodriguez, M. S., Kemp, G. D. & Hay, R. T. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274, 10618–10624 (1999).
Azuma, Y. et al. Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 15, 1825–1827 (2001).
Hay, R. T. SUMO: a history of modification. Mol. Cell 18, 1–12 (2005).
Johnson, E. S. Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 (2004).
Reverter, D. & Lima, C. D. Insights into E3 ligase activity revealed by a SUMO-–RanGAP1–Ubc9–Nup358 complex. Nature 435, 687–692 (2005).
Sampson, D. A., Wang, M. & Matunis, M. J. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21 664–21669 (2001).
Li, S. J. & Hochstrasser, M. A new protease required for cell-cycle progression in yeast. Nature 398, 246–251 (1999).
Mukhopadhyay, D. & Dasso, M. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286–295 (2007).
Di, B. A. et al. The SUMO-specific protease SENP5 is required for cell division. Mol. Cell. Biol. 26, 4489–4498 (2006).
Cheng, J., Kang, X., Zhang, S. & Yeh, E. T. SUMO-specific protease 1 is essential for stabilization of HIF1α during hypoxia. Cell 131, 584–595 (2007).
Vertegaal, A. C. et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics 5, 2298–2310 (2006).
Matafora, V., D'Amato, A., Mori, S., Blasi, F. & Bachi, A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell. Proteomics 8, 2243–2255 (2009).
Bruderer, R. et al. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 12, 142–148 (2011).
Tatham, M. H., Matic, I., Mann, M. & Hay, R. T. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci. Signal. 4, rs4 (2011).
Sohn, S. Y., Bridges, R. G. & Hearing, P. Proteomic analysis of ubiquitin-like posttranslational modifications induced by the adenovirus E4-ORF3 protein. J. Virol. 89, 1744–1755 (2015).
Tu, J. et al. Functional proteomics study reveals SUMOylation of TFII-I is involved in liver cancer cell proliferation. J. Proteome Res. 14, 2385–2397 (2015).
Acknowledgements
The authors apologize to researchers whose work could not be cited owing to space constraints. The authors are grateful for support from the Netherlands Organization for Scientific Research (NWO) and the European Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (box)
Methods for SUMO proteomics (PDF 791 kb)
Supplementary information S2 (table)
A list of all proteins identified to be SUMO target proteins by any of 22 published proteomic studies or low-throughput studies. (XLSX 988 kb)
Supplementary information S3 (table)
A list of all lysines identified to be SUMOylated by any of 11 published proteomic studies or low-throughput studies. (XLSX 2027 kb)
Supplementary information S4 (box)
Methods used for the analysis of SUMO proteomics data (PDF 424 kb)
Supplementary information S5 (table)
A list of all SUMO target proteins identified by SUMOylated lysine, by at least one of 11 published proteomic studies. (XLSX 478 kb)
Supplementary information S6 (box)
Comparison of published and reprocessed sites (PDF 191 kb)
Supplementary information S7 (box)
Technical comparison of SUMO site purification methods (PDF 212 kb)
Supplementary information S8 (box)
Comparison of individual studies (PDF 503 kb)
Supplementary information S9 (box)
Secondary PCA analysis (PDF 917 kb)
Supplementary information S10 (box)
Global effects of stress on the SUMOylated proteome (PDF 269 kb)
Supplementary information S11 (table)
A list of terms, based on Gene Ontology and general keywords, applicable to SUMO target proteins that are globally upregulated or downregulated in SUMOylation in response to proteotoxic stress, as profiled over multiple proteomic studies. (XLSX 32 kb)
Supplementary information S12 (figure)
SUMOylated proteins are functionally interconnected. (PDF 656 kb)
Supplementary information S13 (table)
A list of enriched or depleted terms, based on Gene Ontology, Corum, KEGG, Pfam and general keywords, globally applicable to all SUMO target proteins identified by at least three proteomic studies. (XLSX 245 kb)
Supplementary information S14 (figure)
An unbiased SUMO consensus motif. (PDF 358 kb)
Glossary
- SUMO group modification
-
Simultaneous alteration of a cluster of functionally related proteins by a small ubiquitin-like modifier (SUMO). The cluster contains proteins with SUMO-conjugation sites and SUMO-interaction motifs, which mediate protein–protein interactions.
- KXE-type motifs
-
Small ubiquitin-like modifier (SUMO)ylation consensus motifs consisting of a SUMO-conjugated Lys residue and a Glu residue separated by one amino acid.
- E4 elongase
-
A small ubiquitin-like modifier (SUMO) ligase that extends SUMO polymers.
- Principal component analysis
-
(PCA). An analysis tool used to determine how related or distinct complex data sets are from each other.
- Methyl methanesulfonate
-
An alkylating agent that adds methyl groups to the DNA, thereby causing DNA damage.
- Hydroxyurea
-
A compound that causes DNA replication-dependent damage by inhibiting ribonucleotide reductase, thereby decreasing the production of deoxyribonucleotides.
- [IVL]KXE motif
-
A small ubiquitin-like modifier (SUMO)ylation consensus motif consisting of an inwardly oriented hydrophobic residue, a SUMO-conjugated Lys residue and a Glu residue. This motif is a strong indication for sumoylation and is modified at a relatively high stoichiometry.
- Inverted sumoylation consensus motif
-
(ISCM). A small ubiquitin-like modifier (SUMO)ylation motif in which the acidic residue precedes the sumoylated Lys.
- Tryptic remnant
-
A piece of a protein modification that remains attached to a target protein after digestion of the protein sample by the endoprotease trypsin.
Rights and permissions
About this article
Cite this article
Hendriks, I., Vertegaal, A. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol 17, 581–595 (2016). https://doi.org/10.1038/nrm.2016.81
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2016.81
This article is cited by
-
Hypoxia-driven deSUMOylation of EXOSC10 promotes adaptive changes in the transcriptome profile
Cellular and Molecular Life Sciences (2024)
-
SUMOylation modulates eIF5A activities in both yeast and pancreatic ductal adenocarcinoma cells
Cellular & Molecular Biology Letters (2024)
-
Circ-RAPGEF5 promotes intrahepatic cholangiocarcinoma progression by stabilizing SAE1 to facilitate SUMOylation
Journal of Experimental & Clinical Cancer Research (2023)
-
The emerging roles of SUMOylation in the tumor microenvironment and therapeutic implications
Experimental Hematology & Oncology (2023)
-
BioE3 identifies specific substrates of ubiquitin E3 ligases
Nature Communications (2023)