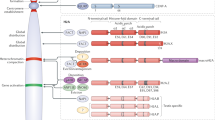
Key Points
-
Chromatin integrity and functionality is governed by the controlled assembly and disassembly of nucleosomes.
-
An elaborate histone chaperone network governs histone provision, chromatin assembly, histone recycling and histone turnover.
-
Histone chaperone networks operate through histone-dependent co-chaperone interactions and direct chaperone–chaperone contacts.
-
The mode of action of histone chaperones is interpreted from structural and biochemical studies of histone–chaperone complexes.
-
Key molecular functions of histone chaperones include the shielding of functional histone interfaces and trapping histones in non-nucleosomal conformations.
-
The integration of histone chaperone function across DNA metabolic processes acts to maintain genome and epigenome integrity.
Abstract
The association of histones with specific chaperone complexes is important for their folding, oligomerization, post-translational modification, nuclear import, stability, assembly and genomic localization. In this way, the chaperoning of soluble histones is a key determinant of histone availability and fate, which affects all chromosomal processes, including gene expression, chromosome segregation and genome replication and repair. Here, we review the distinct structural and functional properties of the expanding network of histone chaperones. We emphasize how chaperones cooperate in the histone chaperone network and via co-chaperone complexes to match histone supply with demand, thereby promoting proper nucleosome assembly and maintaining epigenetic information by recycling modified histones evicted from chromatin.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Laskey, R., Honda, B., Mills, A. & Finch, J. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275, 416–420 (1978). The first description of histone chaperone function elucidated by classical biochemical approaches probing the physicochemical nature of the association between nucleoplasmin and histones.
Venkatesh, S. & Workman, J. L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015).
Alabert, C. & Groth, A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13, 153–167 (2012).
Adam, S., Dabin, J. & Polo, S. E. Chromatin plasticity in response to DNA damage: the shape of things to come. DNA Repair 32, 120–126 (2015).
Annunziato, A. T. Assembling chromatin: the long and winding road. Biochim. Biophys. Acta 1819, 196–210 (2013).
Gurard-Levin, Z. A., Quivy, J. P. & Almouzni, G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 83, 487–517 (2014).
Marzluff, W. F., Wagner, E. J. & Duronio, R. J. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9, 843–854 (2008).
Henikoff, S. & Smith, M. M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 7, a019364 (2015).
O'Sullivan, R. J. et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 21, 167–174 (2014).
Muller, S. & Almouzni, G. A network of players in H3 histone variant deposition and maintenance at centromeres. Biochim. Biophys. Acta 1839, 241–250 (2014).
Heun, P. et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10, 303–315 (2006).
Lacoste, N. et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell 53, 631–644 (2014).
Ishiuchi, T. et al. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat. Struct. Mol. Biol. 22, 662–671 (2015).
Cheloufi, S. et al. The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224 (2015).
Mejlvang, J. et al. New histone supply regulates replication fork speed and PCNA unloading. J. Cell Biol. 204, 29–43 (2014).
Meeks-Wagner, D. & Hartwell, L. H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44, 43–52 (1986).
Gunjan, A. & Verreault, A. A. Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115, 537–549 (2003). Shows that the checkpoint kinase Rad53 monitors the level of soluble histones and that accumulation of excess histones caused by mutation of Rad53 jeopardizes genome stability.
Mattiroli, F., D'Arcy, S. & Luger, K. The right place at the right time: chaperoning core histone variants. EMBO Rep. 16, 1454–1466 (2015).
Andrews, A. J., Chen, X., Zevin, A., Stargell, L. A. & Luger, K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol. Cell 37, 834–842 (2010). Thermodynamic study detailing the role of Nap1 in nucleosome assembly, showing how histone chaperones can buffer interactions between histones and DNA.
Andrews, A. J., Downing, G., Brown, K., Park, Y.-J. & Luger, K. A. Thermodynamic model for Nap1–histone Interactions. J. Biol. Chem. 283, 32412–32418 (2008).
Daganzo, S. M. et al. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13, 2148–2158 (2003).
English, C. M., Adkins, M. W., Carson, J. J., Churchill, M. E. & Tyler, J. K. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006).
Natsume, R. et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 (2007). References 22 and 23 provide the first co-crystal structure of a histone chaperone in complex with its histone cargo, providing a molecular description of how Asf1 chaperones an H3–H4 dimer.
Wang, A. Y. et al. Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc. Natl Acad. Sci. USA 106, 21573–21578 (2009).
Shanle, E. K. et al. Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev. 29, 1795–1800 (2015).
Li, Y. et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell 159, 558–571 (2014).
Liu, H. et al. Structural insights into yeast histone chaperone Hif1: a scaffold protein recruiting protein complexes to core histones. Biochem. J. 462, 465–473 (2014).
Bowman, A. et al. The histone chaperone sNASP binds a conserved peptide motif within the globular core of histone H3 through its TPR repeats. Nucleic Acids Res. 44, 3105–3117 (2015).
Hu, H. et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906 (2011).
Cho, U.-S. S. & Harrison, S. C. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc. Natl Acad. Sci. USA 108, 9367–9371 (2011).
Elsässer, S. et al. DAXX envelops a histone H3.3–H4 dimer for H3.3-specific recognition. Nature 491, 560–565 (2012). Defines the structural specificity of DAXX for H3.3–H4 and describes the sequestration of the H3 α N helix and H4 C terminus in non-nucleosomal conformations. This study uses a novel approach to reconstitute the histone chaperone complex from unfolded components. See also Ref. 32.
Liu, C.-P. et al. Structure of the variant histone H3.3–H4 heterodimer in complex with its chaperone DAXX. Nat. Struct. Mol. Biol. 19, 1287–1292 (2012).
Chen, S. et al. Structure–function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2. Genes Dev. 29, 1326–1340 (2015). Presents the structure of human SPT2 bound to a H3–H4 tetramer and shows that histone-binding mutants of Spt2 in yeast phenocopy Spt2 deletion in failing to suppress cryptic transcription.
Huang, H. et al. A unique binding mode enables MCM2 to chaperone histones H3–H4 at replication forks. Nat. Struct. Mol. Biol. 22, 618–626 (2015). Provides the crystal structure of MCM2 with a H3–H4 tetramer and shows that MCM2 in vivo chaperones H3–H4 alone and as part of the replicative helicase. Also provides the first structure of a co-chaperone complex, the MCM2–H3–H4–ASF1 complex.
Richet, N. et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 43, 1905–1917 (2015).
Belotserkovskaya, R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003). Describes the histone chaperone function of the FACT complex and its ability to promote H2A–H2B eviction and transcription through chromatin.
Tsunaka, Y., Fujiwara, Y., Oyama, T., Hirose, S. & Morikawa, K. Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev. 30, 673–686 (2016). Elucidates structurally how SPT16 can bind a H3–H4 tetramer and provides insights into the mechanism of nucleosome disruption by the FACT complex.
Foltman, M. et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 3, 892–904 (2013).
Liu, Y. et al. Structural analysis of Rtt106p reveals a DNA binding role required for heterochromatin silencing. J. Biol. Chem. 285, 4251–4262 (2010).
VanDemark, A. et al. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 22, 363–374 (2006).
Hondele, M. et al. Structural basis of histone H2A–H2B recognition by the essential chaperone FACT. Nature 499, 111–114 (2013).
Li, Q. et al. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134, 244–255 (2008). Shows that H3K56 acetylation promotes binding of histones to Rtt106 and the CAF1 complex and determines that the tandem pleckstrin homology domain of Rtt106 has specificity for H3K56ac.
Su, D. et al. Structural basis for recognition of H3K56-acetylated histone H3–H4 by the chaperone Rtt106. Nature 483, 104–107 (2012).
Fazly, A. et al. Histone chaperone Rtt106 promotes nucleosome formation using (H3–H4)2 tetramers. J. Biol. Chem. 287, 10753–10760 (2012).
Stuwe, T. et al. The FACT Spt16 “peptidase” domain is a histone H3–H4 binding module. Proc. Natl Acad. Sci. USA 105, 8884–8889 (2008).
Kemble, D. J., McCullough, L. L., Whitby, F. G., Formosa, T. & Hill, C. P. FACT disrupts nucleosome structure by binding H2A–H2B with conserved peptide motifs. Mol. Cell 60, 294–306 (2015).
Formosa, T. The role of FACT in making and breaking nucleosomes. Biochim. Biophys. Acta 1819, 247–255 (2012).
Selth, L. & Svejstrup, J. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 282, 12358–12362 (2007).
Park, Y.-J. & Luger, K. The structure of nucleosome assembly protein 1. Proc. Natl Acad. Sci. USA 103, 1248–1253 (2006).
Muto, S. et al. Relationship between the structure of SET/TAF-Iβ/INHAT and its histone chaperone activity. Proc. Natl Acad. Sci. USA 104, 4285–4290 (2007).
Hammond, C. M. et al. The histone chaperone Vps75 forms multiple oligomeric assemblies capable of mediating exchange between histone H3–H4 tetramers and Asf1–H3–H4 complexes. Nucleic Acids Res. 44, 6157–6172 (2016). Describes how Vps75 forms a co-chaperone complex with Asf1 and histones, and provides the first description of a self-chaperoning mechanism for histone chaperones by demonstrating that the histone-binding surface is shielded in the inactive Vps75 tetramer.
Aguilar-Gurrieri, C. et al. Structural evidence for Nap1-dependent H2A–H2B deposition and nucleosome assembly. EMBO J. 35, 1465–1482 (2016). Provides the crystal structure of Nap1 with H2A–H2B, showing how Nap1 can buffer H2A–H2B interactions with DNA, and (together with reference 51) sets the precedence for Nap1-like proteins binding directly to a histone dimer.
Bowman, A. et al. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol. Cell 41, 398–408 (2011).
D'Arcy, S. & Luger, K. Understanding histone acetyltransferase Rtt109 structure and function: how many chaperones does it take? Curr. Opin. Struct. Biol. 21, 728–734 (2011).
Zlatanova, J., Seebart, C. & Tomschik, M. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 21, 1294–1310 (2007).
Bowman, A. et al. The histone chaperones Vps75 and Nap1 form ring-like, tetrameric structures in solution. Nucleic Acids Res. 42, 6038–6051 (2014).
Newman, E. R. et al. Large multimeric assemblies of nucleosome assembly protein and histones revealed by small-angle X-ray scattering and electron microscopy. J. Biol. Chem. 287, 26657–26665 (2012).
Tang, Y. et al. Structure of the Rtt109–AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation. Structure 19, 221–231 (2011).
Kolonko, E. M. et al. Catalytic activation of histone acetyltransferase Rtt109 by a histone chaperone. Proc. Natl Acad. Sci. USA 107, 20275–20280 (2010).
Su, D. et al. Structure and histone binding properties of the Vps75–Rtt109 chaperone–lysine acetyltransferase complex. J. Biol. Chem. 286, 15625–15629 (2011).
Dutta, S. et al. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8, 841–853 (2001).
Namboodiri, V. M., Akey, I. V. V., Schmidt-Zachmann, M. S., Head, J. F. & Akey, C. W. The structure and function of Xenopus NO38-core, a histone chaperone in the nucleolus. Structure 12, 2149–2160 (2004).
Fernández-Rivero, N. et al. A quantitative characterization of nucleoplasmin/histone complexes reveals chaperone versatility. Sci. Rep. 6, 32114 (2016).
Ramos, I. et al. Nucleoplasmin binds histone H2A–H2B dimers through its distal face. J. Biol. Chem. 285, 33771–33778 (2010).
Luger, K., Mäder, A., Richmond, R., Sargent, D. & Richmond, T. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997). This seminal work provides a detailed molecular description of the nucleosome that has inspired many young scientists to enter the chromatin field.
Arents, G., Burlingame, R. W., Wang, B. C., Love, W. E. & Moudrianakis, E. N. The nucleosomal core histone octamer at 3.1 Å resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl Acad. Sci. USA 88, 10148–10152 (1991).
Hong, J. et al. The catalytic subunit of the SWR1 remodeler is a histone chaperone for the H2A.Z–H2B dimer. Mol. Cell 53, 498–505 (2014).
Mao, Z. et al. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell Res. 24, 389–399 (2014).
Obri, A. et al. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–653 (2014). References 68 and 69 identify the first H2AZ-specific histone chaperone in mammalian cells, ANP32E, providing structural details of the interaction and describing a specific role in H2AZ–H2B eviction.
Liang, X. et al. Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1. Nat. Struct. Mol. Biol. 23, 317–323 (2016).
Latrick, C. M. et al. Molecular basis and specificity of H2A.Z–H2B recognition and deposition by the histone chaperone YL1. Nat. Struct. Mol. Biol. 23, 309–316 (2016).
Donham, D., Scorgie, J. & Churchill, M. The activity of the histone chaperone yeast Asf1 in the assembly and disassembly of histone H3/H4–DNA complexes. Nucleic Acids Res. 39, 5449–5458 (2011).
Chavez, M. S. et al. The conformational flexibility of the C-terminus of histone H4 promotes histone octamer and nucleosome stability and yeast viability. Epigenetics Chromatin 5, 5 (2012).
Song, J.-J., Garlick, J. D. & Kingston, R. E. Structural basis of histone H4 recognition by p55. Genes Dev. 22, 1313–1318 (2008).
Murzina, N. V. et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085 (2008).
Zhang, W. et al. Structural plasticity of histones H3–H4 facilitates their allosteric exchange between RbAp48 and ASF1. Nat. Struct. Mol. Biol. 20, 29–35 (2013).
Schmitges, F. W. et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 (2011).
Li, Y. et al. Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex. Genes Dev. 28, 1217–1227 (2014).
An, S., Kim, H. & Cho, U.-S. Mis16 independently recognizes histone H4 and the CENP-ACnp1-specific chaperone Scm3sp. J. Mol. Biol. 427, 3230–3240 (2015).
Furuyama, T., Dalal, Y. & Henikoff, S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc. Natl Acad. Sci. USA 103, 6172–6177 (2006).
Wang, H., Wang, M., Yang, N. & Xu, R.-M. Structure of the quaternary complex of histone H3–H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell 6, 693–697 (2015).
Zhou, Z. et al. NMR structure of chaperone Chz1 complexed with histones H2A.Z–H2B. Nat. Struct. Mol. Biol. 15, 868–869 (2008).
DeNizio, J. E., Elsässer, S. J. & Black, B. E. DAXX co-folds with H3.3/H4 using high local stability conferred by the H3.3 variant recognition residues. Nucleic Acids Res. 42, 4318–4331 (2014).
Saredi, G. et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex. Nature 534, 714–718 (2016). Reveals a dual histone chaperone and reader function of TONSL, binding H4 tails that are unmethylated at K20 on soluble and nucleosomal histones. H4K20me0 is specific to newly synthesized histones, allowing recruitment of the TONSL–MMS22L repair complex to post-replicative chromatin.
Campos, E. I. et al. Analysis of the histone H3. interactome: a suitable chaperone for the right event. Mol. Cell 60, 697–709 (2015).
Kalashnikova, A. A., Porter-Goff, M. E., Muthurajan, U. M., Luger, K. & Hansen, J. C. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface 10, 20121022 (2013).
Jasencakova, Z. et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 37, 736–743 (2010). This mass spectrometry analysis of modifications on ASF1-bound histones demonstrates that replication stress induces K9me1 accumulation on H3 and impairs recycling of old H3–H4.
Groth, A. et al. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17, 301–311 (2005).
Groth, A. et al. Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 (2007). Identifies the H3–H4-dependent interaction between ASF1 and the MCM2–7 helicase and reveals the requirement of ASF1 for DNA unwinding, coupling histone chaperone function to DNA replication.
Campos, E. et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 17, 1343–1351 (2010). Characterizes distinct H3.1–H4 chaperone complexes present in the cytosolic fraction and identifies HSC70 and HSP90 as upstream chaperones.
Haigney, A., Ricketts, M. D. & Marmorstein, R. Dissecting the molecular roles of histone chaperones in histone acetylation by type B histone acetyltransferases (HAT-B). J. Biol. Chem. 290, 30648–30657 (2015).
D'Andrea, L. D. & Regan, L. TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662 (2003).
Daniel Ricketts, M. et al. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6, 7711 (2015).
Malay, A. D., Umehara, T., Matsubara-Malay, K., Padmanabhan, B. & Yokoyama, S. Crystal structures of fission yeast histone chaperone Asf1 complexed with the Hip1 B-domain or the Cac2 C terminus. J. Biol. Chem. 283, 14022–14031 (2008).
Tang, Y. et al. Structure of a human ASF1a–HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13, 921–929 (2006).
Ask, K. et al. Codanin-1, mutated in the anaemic disease CDAI, regulates Asf1 function in S-phase histone supply. EMBO J. 31, 2013–2023 (2012).
Jiao, Y. et al. Surprising complexity of the Asf1 histone chaperone–Rad53 kinase interaction. Proc. Natl Acad. Sci. USA 109, 2866–2871 (2012).
Kurat, C. F. et al. Regulation of histone gene transcription in yeast. Cell. Mol. Life Sci. 71, 599–613 (2014).
Singh, R. K., Kabbaj, M. H., Paik, J. & Gunjan, A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 11, 925–933 (2009).
Nelson, D. M. et al. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 22, 7459–7472 (2002).
Moshkin, Y. M. et al. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 16, 2621–2626 (2002).
Klimovskaia, I. M. et al. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nat. Commun. 5, 3394 (2014).
Sillje, H. H. & Nigg, E. A. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11, 1068–1073 (2001).
Gerard, A. et al. The replication kinase Cdc7-Dbf4 promotes the interaction of the p150 subunit of chromatin assembly factor 1 with proliferating cell nuclear antigen. EMBO Rep. 7, 817–823 (2006).
Muller, S. et al. Phosphorylation and DNA binding of HJURP determine its centromeric recruitment and function in CenH3CENP-A loading. Cell Rep. 8, 190–203 (2014).
Luk, E. et al. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell 25, 357–368 (2007). Identifies the first H2AZ-specific histone chaperone in yeast, Chz1, and defines the H2AZ interaction motif of Chz1.
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004). Identifies distinct complexes of H3.1 and H3.3 with CAF1 and HIRA complexes, respectively, and demonstrates that soluble H3–H4 are mainly found as dimers.
Drane, P., Ouararhni, K., Depaux, A., Shuaib, M. & Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 (2010).
Alvarez, F. et al. Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 286, 17714–17721 (2011).
Parthun, M. R. Histone acetyltransferase 1: more than just an enzyme? Biochim. Biophys. Acta 1819, 256–263 (2012).
Cook, A. J., Gurard-Levin, Z. A., Vassias, I. & Almouzni, G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3–H4 in the histone supply chain. Mol. Cell 44, 918–927 (2011). References 90 and 111 identify NASP as a H3–H4-specific chaperone, required for storage of H3–H4 and to prevent its degradation.
Poveda, A. et al. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J. Biol. Chem. 279, 16033–16043 (2004).
Green, E. M. et al. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15, 2044–2049 (2005).
Tyler, J. K. et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 (1999). Identifies Asf1 as a H3–H4 chaperone that cooperates with the CAF1 complex in replication-coupled chromatin assembly.
Ray-Gallet, D. et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 (2002).
Mello, J. A. et al. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3, 329–334 (2002).
Tyler, J. K. et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21, 6574–6584 (2001).
Benson, L. J. et al. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 281, 9287–9296 (2006).
Abascal, F. et al. Subfunctionalization via adaptive evolution influenced by genomic context: the case of histone chaperones ASF1a and ASF1b. Mol. Biol. Evol. 30, 1853–1866 (2013).
Hayashi, R. et al. Transcriptional regulation of human chromatin assembly factor ASF1. DNA Cell Biol. 26, 91–99 (2007).
Corpet, A. et al. Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 30, 480–493 (2011).
Quivy, J. P., Gerard, A., Cook, A. J., Roche, D. & Almouzni, G. The HP1–p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat. Struct. Mol. Biol. 15, 972–979 (2008).
Hoek, M. & Stillman, B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl Acad. Sci. USA 100, 12183–12188 (2003).
Nabatiyan, A., Szuts, D. & Krude, T. Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells. Mol. Cell. Biol. 26, 1839–1849 (2006).
Piwko, W., Buser, R. & Peter, M. Rescuing stalled replication forks: MMS22L–TONSL, a novel complex for DNA replication fork repair in human cells. Cell Cycle 10, 1703–1705 (2011).
Straube, K., Blackwell, J. S. Jr & Pemberton, L. F. Nap1 and Chz1 have separate Htz1 nuclear import and assembly functions. Traffic 11, 185–197 (2010).
Cai, Y. et al. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280, 13665–13670 (2005).
Parthun, M. R. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene 26, 5319–5328 (2007).
Loyola, A., Bonaldi, T., Roche, D., Imhof, A. & Almouzni, G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24, 309–316 (2006). This mass spectrometry analysis of histone modifications on soluble and nucleosomal H3.1–H4 and H3.3–H4 complexes identifies H3K9me1 as a predeposition mark that is important for establishing heterochromatin.
Masumoto, H., Hawke, D., Kobayashi, R. & Verreault, A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 (2005). Identifies H3K56 acetylation as a highly abundant mark on new soluble histones in yeast, defines the cell cycle dynamics of the mark and shows that it is required for genome stability.
Chen, C.-C. et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134, 231–243 (2008).
Alabert, C. et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29, 585–590 (2015).
Xu, F., Zhang, K. & Grunstein, M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121, 375–385 (2005).
Han, J., Zhang, H., Wang, Z., Zhou, H. & Zhang, Z. A. Cul4 E3 ubiquitin ligase regulates histone hand-off during nucleosome assembly. Cell 155, 817–829 (2013).
Niikura, Y. et al. CENP-A K124 ubiquitylation is required for CENP-A deposition at the centromere. Dev. Cell 32, 589–603 (2015).
Rivera, C. et al. Methylation of histone H3 lysine 9 occurs during translation. Nucleic Acids Res. 43, 9097–9106 (2015).
Loyola, A. et al. The HP1α–CAF1–SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10, 769–775 (2009).
Shibahara, K. & Stillman, B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999). Seminal paper showing that the CAF1 complex is recruited to replication forks through interaction with PCNA to mediate replication-coupled chromatin assembly.
Smith, D. J. & Whitehouse, I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438 (2012).
Moggs, J. G. et al. A CAF-1–PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol. Cell. Biol. 20, 1206–1218 (2000).
Polo, S. E., Roche, D. & Almouzni, G. New histone incorporation marks sites of UV repair in human cells. Cell 127, 481–493 (2006).
Li, X. & Tyler, J. K. Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. eLife 5, e15129 (2016).
Brachet, E., Beneut, C., Serrentino, M. E. & Borde, V. The CAF-1 and Hir histone chaperones associate with sites of meiotic double-strand breaks in budding yeast. PLoS ONE 10, e0125965 (2015).
Huang, S. et al. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc. Natl Acad. Sci. USA 102, 13410–13415 (2005).
Yang, J. et al. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 14, 1128–1141 (2016).
Huang, C. & Zhu, B. H3.3 turnover: a mechanism to poise chromatin for transcription, or a response to open chromatin? Bioessays 36, 579–584 (2014).
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010). References 108 and 147 identify DAXX–ATRX as a H3.3-specific chaperone complex. DAXX–ATRX affinity tags endogenous H3.3 for genome-wide profiling, showing HIRA-dependent incorporation in genomic regions and ATRX-dependent incorporation in telomeres.
Elsasser, S. J., Noh, K. M., Diaz, N., Allis, C. D. & Banaszynski, L. A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244 (2015).
Lewis, P. W., Elsaesser, S. J., Noh, K. M., Stadler, S. C. & Allis, C. D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA 107, 14075–14080 (2010).
Banaszynski, L. A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Ray-Gallet, D. et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44, 928–941 (2011).
Pchelintsev, N. A. et al. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 3, 1012–1019 (2013).
Soni, S., Pchelintsev, N., Adams, P. D. & Bieker, J. J. Transcription factor EKLF (KLF1) recruitment of the histone chaperone HIRA is essential for β-globin gene expression. Proc. Natl Acad. Sci. USA 111, 13337–13342 (2014).
Adam, S., Polo, S. E. & Almouzni, G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 155, 94–106 (2013).
Iwase, S. et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat. Struct. Mol. Biol. 18, 769–776 (2011).
Dhayalan, A. et al. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum. Mol. Genet. 20, 2195–2203 (2011).
Eustermann, S. et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 18, 777–782 (2011).
Xiao, H. et al. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol. Cell 43, 369–380 (2011).
Stoler, S. et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl Acad. Sci. USA 104, 10571–10576 (2007).
Camahort, R. et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26, 853–865 (2007).
Shivaraju, M., Camahort, R., Mattingly, M. & Gerton, J. L. Scm3 is a centromeric nucleosome assembly factor. J. Biol. Chem. 286, 12016–12023 (2011).
Mizuguchi, G., Xiao, H., Wisniewski, J., Smith, M. M. & Wu, C. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell 129, 1153–1164 (2007).
Wisniewski, J. et al. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. eLife 3, e02203 (2014).
Lando, D. et al. Quantitative single-molecule microscopy reveals that CENP-ACnp1 deposition occurs during G2 in fission yeast. Open Biol. 2, 120078 (2012).
Jansen, L. E., Black, B. E., Foltz, D. R. & Cleveland, D. W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805 (2007). Introduces SNAP-tag pulse-labelling technology to track old and new CENP-A, showing efficient recycling of old CENP-A in S phase of the cell cycle complemented by de novo deposition of CENP-A in G1 phase.
Dunleavy, E. M. et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009). References 166 and 167 identify HJURP as a CENP-A-specific chaperone required for deposition of new CENP-A in early G1 phase.
Foltz, D. R. et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009).
Pidoux, A. L. et al. Fission yeast Scm3: a CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33, 299–311 (2009).
Westhorpe, F. G., Fuller, C. J. & Straight, A. F. A cell-free CENP-A assembly system defines the chromatin requirements for centromere maintenance. J. Cell Biol. 209, 789–801 (2015).
Foltz, D. R. et al. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469 (2006).
Wu, W. H. et al. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat. Struct. Mol. Biol. 12, 1064–1071 (2005).
Wu, W. H. et al. N terminus of Swr1 binds to histone H2AZ and provides a platform for subunit assembly in the chromatin remodeling complex. J. Biol. Chem. 284, 6200–6207 (2009).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004). Establishes an elegant in vitro assay to show that SWR1 mediates the ATP-dependent exchange of canonical H2A with the H2A. Z variant, paving the way for an in-depth understanding of SWR1 function.
Luk, E. et al. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736 (2010).
Ranjan, A. et al. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell 154, 1232–1245 (2013).
Yen, K., Vinayachandran, V. & Pugh, B. F. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154, 1246–1256 (2013).
Liu, W. H., Roemer, S. C., Port, A. M. & Churchill, M. E. CAF-1-induced oligomerization of histones H3/H4 and mutually exclusive interactions with Asf1 guide H3/H4 transitions among histone chaperones and DNA. Nucleic Acids Res. 40, 11229–11239 (2012).
Winkler, D., Zhou, H., Dar, M., Zhang, Z. & Luger, K. Yeast CAF-1 assembles histone (H3–H4)2 tetramers prior to DNA deposition. Nucleic Acids Res. 40, 10139–10149 (2012).
Dechassa, M. L., Wyns, K. & Luger, K. Scm3 deposits a (Cse4–H4)2 tetramer onto DNA through a Cse4–H4 dimer intermediate. Nucleic Acids Res. 42, 5532–5542 (2014).
Zasadzin´ska, E., Barnhart-Dailey, M. C., Kuich, H. P. J. L. & Foltz, D. R. Dimerization of the CENP-A assembly factor HJURP is required for centromeric nucleosome deposition. EMBO J. 32, 2113–2124 (2013).
Ahmad, K. & Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 (2002).
Black, B. E. et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 25, 309–322 (2007).
Adkins, M. W., Howar, S. R. & Tyler, J. K. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14, 657–666 (2004).
Korber, P. et al. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 281, 5539–5545 (2006).
Schwabish, M. A. & Struhl, K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22, 415–422 (2006).
Kuryan, B. G. et al. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc. Natl Acad. Sci. USA 109, 1931–1936 (2012).
Chen, X. et al. The histone chaperone Nap1 is a major regulator of histone H2A–H2B dynamics at the inducible GAL locus. Mol. Cell. Biol. 36, 1287–1296 (2016).
Jamai, A., Puglisi, A. & Strubin, M. Histone chaperone spt16 promotes redeposition of the original H3–H4 histones evicted by elongating RNA polymerase. Mol. Cell 35, 377–383 (2009).
Voth, W. P. et al. A role for FACT in repopulation of nucleosomes at inducible genes. PLoS ONE 9, e84092 (2014).
Schwabish, M. A. & Struhl, K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24, 10111–10117 (2004).
Kaplan, C. D., Laprade, L. & Winston, F. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301, 1096–1099 (2003).
Xue, Y.-M. M. et al. Histone chaperones Nap1 and Vps75 regulate histone acetylation during transcription elongation. Mol. Cell. Biol. 33, 1645–1656 (2013).
Bortvin, A. & Winston, F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272, 1473–1476 (1996).
Osakabe, A. et al. Vertebrate Spt2 is a novel nucleolar histone chaperone that assists in ribosomal DNA transcription. J. Cell Sci. 126, 1323–1332 (2013).
Nourani, A., Robert, F. & Winston, F. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 1496–1509 (2006).
Selth, L. et al. An rtt109-independent role for vps75 in transcription-associated nucleosome dynamics. Mol. Cell. Biol. 29, 4220–4234 (2009).
Kaplan, T. et al. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4, e1000270 (2008).
Venkatesh, S. et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 (2012).
Pavri, R. et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125, 703–717 (2006).
Fleming, A. B., Kao, C. F., Hillyer, C., Pikaart, M. & Osley, M. A. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell 31, 57–66 (2008).
Trujillo, K. M. & Osley, M. A. A role for H2B ubiquitylation in DNA replication. Mol. Cell 48, 734–746 (2012).
Annunziato, A. T. Split decision: what happens to nucleosomes during DNA replication? J. Biol. Chem. 280, 12065–12068 (2005).
Alabert, C. et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–293 (2014).
Gambus, A. et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 (2006).
Tan, B. C., Chien, C. T., Hirose, S. & Lee, S. C. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 25, 3975–3985 (2006).
McCullough, L., Connell, Z., Petersen, C. & Formosa, T. The abundant histone chaperones Spt6 and FACT collaborate to assemble, inspect, and maintain chromatin structure in Saccharomyces cerevisiae. Genetics 201, 1031–1045 (2015).
Nekrasov, M. et al. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nat. Struct. Mol. Biol. 19, 1076–1083 (2012).
Xu, M. et al. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–98 (2010). This mass spectrometry analysis of tagged H3.1 and H3.3 mononucleosomes shows that new and old H3.1–H4 dimers (unlike H3.3–H4 dimers) do not mix but can be mixed with new and old H2A–H2B.
Ishimi, Y., Komamura, Y., You, Z. & Kimura, H. Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem. 273, 8369–8375 (1998).
Ishimi, Y., Komamura-Kohno, Y., Arai, K. & Masai, H. Biochemical activities associated with mouse Mcm2 protein. J. Biol. Chem. 276, 42744–42752 (2001).
Sun, J. et al. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 22, 976–982 (2015).
Wittmeyer, J., Joss, L. & Formosa, T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase α. Biochemistry 38, 8961–8971 (1999).
Bowman, A., Ward, R., El-Mkami, H., Owen-Hughes, T. & Norman, D. Probing the (H3–H4)2 histone tetramer structure using pulsed EPR spectroscopy combined with site-directed spin labelling. Nucleic Acids Res. 38, 695–707 (2010). Describes the structure of the H3–H4 tetramer and highlights the dynamic nature of the H3 α N helix in the absence of H2A–H2B.
Acknowledgements
The authors apologize for the many studies in the histone chaperone field that were unable to be cited owing to space restrictions. The authors thank C. Alabert, A. Bowman and Z. Jasencakova for useful comments on the manuscript and for help with figure design. D.J.P. is supported by funds from the Leukemia and Lymphoma Society and STARR Foundation and by the Memorial Sloan-Kettering Cancer Core Grant (P30 CA008748). A.G. is an EMBO Young Investigator and her research is supported by the European Research Council (ERC StG, no. 281765), the Danish National Research Foundation to the Center for Epigenetics (DNRF82), the Danish Cancer Society, the Danish Medical Research Council, the Novo Nordisk Foundation and the Lundbeck Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare competing financial interests: C.M.H., H.H., D.J.P. and A.G. are named inventors on a patent application covering the discoveries in Saredi et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex. Nature 534, 714–718 (2016).
Related links
Glossary
- Histone chaperones
-
Defined here as proteins that handle non-nucleosomal histones in vivo and mediate the assembly of nucleosomes from isolated histones and DNA in vitro.
- Histone storage
-
The sequestration of histones in the soluble fraction of the cell that prevents their degradation.
- Histone turnover
-
The eviction of nucleosomal histones, followed by deposition of new histones at the same genomic loci.
- Histone chaperone network
-
The integration of histone chaperone functions to support histone dynamics across various cellular processes.
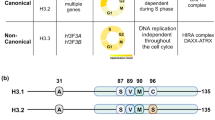
- Canonical histones
-
Core histone subtypes (H3.1, H3.2, H4, H2A and H2B) that are expressed in S phase of the cell cycle and mainly incorporated into nucleosomes in a DNA replication-dependent manner.
- Replacement variants
-
Histone subtypes (such as H3.3, CENP-A, and H2A.Z) incorporated into nucleosomes via DNA replication-independent pathways and for which expression is not restricted to S phase.
- De novo histone deposition
-
Incorporation of newly synthesized histones into chromatin.
- Co-chaperone
-
Here defined as a complex containing two or more histone chaperones brought together in a histone-dependent manner.
- Epigenetic plasticity
-
Heritable information other than DNA sequence that maintains cellular traits while also being subject to change without said changes being permanent.
- H3K56ac
-
A mark of newly synthesized H3–H4 in yeast, catalysed by Rtt109 in an Asf1-dependent manner, that promotes replication-dependent histone deposition.
- Histone recycling
-
Re-deposition of histones evicted from chromatin by cellular processes that require access to the DNA template.
- Dyad DNA
-
The dyad position locates the pseudo axis of symmetry, which coincides with the central base pair (or pairs) of nucleosomal DNA and the H3–H4 tetramerization interface, around which the nucleosome can be rotated 180° and map back onto itself.
- Tetrasome
-
Thought to be the first assembly intermediate during nucleosome assembly, the tetrasome is the product of the deposition of a H3–H4 tetramer on DNA.
- RBAP46 and RBAP48
-
Histone chaperone homologues that are almost identical and seem to be interchangeable in most of their chromatin-modifying complexes, apart from HAT1 (RBAP46) and CAF1 (RBAP48).
- Histone reader
-
A protein that binds to histones in a post-translational modification-dependent manner.
- H4K20me0
-
Histone H4 unmethylated at lysine 20 (H4K20me0); a signature of newly synthesized histones that marks post-replicative chromatin until G2/M phase of the cell cycle, when H4K20 methylation is established on those new histones.
- Soluble histones
-
Non-nucleosomal histones.
- H4K5acK12ac
-
Highly conserved diacetylation mark, catalysed by RBAP46–HAT1, that marks newly synthesized histone H4 before deposition.
- Histone exchange
-
The replacement of nucleosomal histones with the corresponding canonical histones (H2A–H2B, H3–H4) or replacement variants (H2A.Z–H2B, H3.3–H4).
- Hexasome
-
A nucleosome intermediate generated by either the loss of one H2A–H2B dimer from the nucleosome or the addition of one H2A–H2B dimer to the H3–H4 tetrasome.
Rights and permissions
About this article
Cite this article
Hammond, C., Strømme, C., Huang, H. et al. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol 18, 141–158 (2017). https://doi.org/10.1038/nrm.2016.159
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrm.2016.159
This article is cited by
-
HIRA vs. DAXX: the two axes shaping the histone H3.3 landscape
Experimental & Molecular Medicine (2024)
-
DNAJC9 prevents CENP-A mislocalization and chromosomal instability by maintaining the fidelity of histone supply chains
The EMBO Journal (2024)
-
Elimination of antibiotic-resistant bacteria and resistance genes by earthworms during vermifiltration treatment of excess sludge
Environmental Science and Pollution Research (2024)
-
The histone chaperone SPT2 regulates chromatin structure and function in Metazoa
Nature Structural & Molecular Biology (2024)
-
FACT regulates pluripotency through proximal and distal regulation of gene expression in murine embryonic stem cells
BMC Biology (2023)