Key Points
-
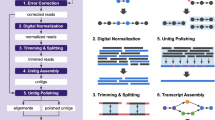
The protocols used for library construction, sequencing and data pre-processing can have a great impact on the quality of an assembled transcriptome and the accuracy of gene expression quantification.
-
Before starting an RNA sequencing (RNA-seq) experiment, one should carefully consider using protocols that are strand-specific, that remove ribosomal RNA and that do not require PCR amplification of the template.
-
Strand-specific RNA-seq protocols are important for correctly assembling overlapping transcripts, especially for compact genomes.
-
The reference-based, or ab initio, assembly strategy requires a reference genome and uses much fewer computing resources than the de novo strategy. However, the quality of the genome and the ability of the short-read aligner to align reads across introns will directly influence the accuracy of the assembled transcripts when using the reference-based strategy.
-
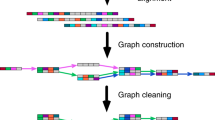
The de novo assembly strategy does not use a reference genome but instead uses a De Bruijn graph to represent overlaps between sequences and assemble transcripts. Most de novo approaches require significant computing resources: random access memory (RAM) is the typical limitation. However, de novo assemblers can assemble trans-spliced genes and novel transcripts that are not present in the genome assembly.
-
To take full advantage of the current assembly strategies, a combined assembly approach should be considered that leverages the strengths of reference-based and de novo assembly strategies.
-
Most transcriptome assemblers are still being developed, and the results from these programs should be evaluated using unbiased quantitative metrics.
-
Transcriptome assembly involves an informatics approach to solve an experimental limitation. As sequencing strategies continually improve, it may no longer be necessary in the near future to assemble transcriptomes, as the read length will be longer than any individual transcript.
Abstract
Transcriptomics studies often rely on partial reference transcriptomes that fail to capture the full catalogue of transcripts and their variations. Recent advances in sequencing technologies and assembly algorithms have facilitated the reconstruction of the entire transcriptome by deep RNA sequencing (RNA-seq), even without a reference genome. However, transcriptome assembly from billions of RNA-seq reads, which are often very short, poses a significant informatics challenge. This Review summarizes the recent developments in transcriptome assembly approaches — reference-based, de novo and combined strategies — along with some perspectives on transcriptome assembly in the near future.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ozsolak, F. & Milos, P. M. RNA sequencing: advances, challenges and opportunities. Nature Rev. Genet. 12, 87–98 (2011). This Review provides a good, up-to-date summary of the RNA-seq experimental protocol and its usefulness in addressing important biological questions.
Wang, Z., Gerstein, M. & Snyder, M. RNA-seq: a revolutionary tool for transcriptomics. Nature Rev. Genet. 10, 57–63 (2009).
Marguerat, S. & Bahler, J. RNA-seq: from technology to biology. Cell. Mol. Life Sci. 67, 569–579 (2010).
Wilhelm, B. T. & Landry, J. R. RNA-seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48, 249–257 (2009).
Metzker, M. L. Sequencing technologies — the next generation. Nature Rev. Genet. 11, 31–46 (2010). This Review provides a good introduction to NGS technologies and the analysis challenges that they pose.
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Simpson, J. T. et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 19, 1117–1123 (2009).
Butler, J. et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 18, 810–820 (2008).
Paszkiewicz, K. & Studholme, D. J. De novo assembly of short sequence reads. Brief. Bioinform. 11, 457–472 (2010).
Miller, J. R., Koren, S. & Sutton, G. Assembly algorithms for next-generation sequencing data. Genomics 95, 315–327 (2010). This paper provides a good introduction to the current algorithms used in next-generation genome assembly and the challenges posed by these approaches.
Makalowska, I., Lin, C. F. & Makalowski, W. Overlapping genes in vertebrate genomes. Comput. Biol. Chem. 29, 1–12 (2005).
Normark, S. et al. Overlapping genes. Annu. Rev. Genet. 17, 499–525 (1983).
Johnson, Z. I. & Chisholm, S. W. Properties of overlapping genes are conserved across microbial genomes. Genome Res. 14, 2268–2272 (2004).
Fukuda, Y., Washio, T. & Tomita, M. Comparative study of overlapping genes in the genomes of Mycoplasma genitalium and Mycoplasma pneumoniae. Nucleic Acids Res. 27, 1847–1853 (1999).
Martin, J. et al. Rnnotator: an automated de novo transcriptome assembly pipeline from stranded RNA-seq reads. BMC Genomics 11, 663 (2010). This paper describes the first de novo transcriptome assembler to automate the use of several k-mers for assembly. It also provides a good overview of methods used for the pre- and post-processing of de novo transcriptome assemblies.
Guttman, M. et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nature Biotech. 28, 503–510 (2010). This paper introduces the Scripture algorithm, which was one of the first reference-based assemblers that effectively tackled the assembly of alternative isoforms using NGS data.
Denoeud, F. et al. Annotating genomes with massive-scale RNA sequencing. Genome Biol. 9, R175 (2008).
Robertson, G. et al. De novo assembly and analysis of RNA-seq data. Nature Methods 7, 909–912 (2010).
Surget-Groba, Y. & Montoya-Burgos, J. I. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 20, 1432–1440 (2010).
Trapnell, C. et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotech. 28, 511–515 (2010). The Cufflinks algorithm is introduced in this paper, which, like the Scripture algorithm described in reference 16, was one of the first reference-based assemblers that effectively tackled the assembly of alternative isoforms using NGS data.
Birol, I. et al. De novo transcriptome assembly with ABySS. Bioinformatics 25, 2872–2877 (2009).
Crawford, J. E. et al. De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-seq technology. PLoS ONE 5, e14202 (2010).
Garg, R., Patel, R. K., Tyagi, A. K. & Jain, M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 18, 53–63 (2011).
Yassour, M. et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc. Natl Acad. Sci. USA 106, 3264–3269 (2009).
Adamidi, C. et al. De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res. 21, 1193–1200 (2011).
Katz, Y., Wang, E. T., Airoldi, E. M. & Burge, C. B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature Methods 7, 1009–1015 (2010).
Levin, J. Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nature Methods 7, 709–715 (2010). This paper provides an excellent comparison of different RNA-seq protocols and how they affect the quantification of expression levels.
He, S. et al. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nature Methods 7, 807–812 (2010).
Chen, Z. & Duan, X. Ribosomal RNA depletion for massively parallel bacterial RNA-sequencing applications. Methods Mol. Biol. 733, 93–103 (2011).
Christodoulou, D. C., Gorham, J. M., Herman, D. S. & Seidman, J. G. Construction of normalized RNA-seq libraries for next-generation sequencing using the crab duplex-specific nuclease. Curr. Protoc. Mol. Biol. 1 Apr 2011 (doi:10.1002/0471142727.mb0412s94).
Kozarewa, I. et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nature Methods 6, 291–295 (2009).
Mamanova, L. et al. FRT-seq: amplification-free, strand-specific transcriptome sequencing. Nature Methods 7, 130–132 (2010).
Sam, L. T. et al. A comparison of single molecule and amplification based sequencing of cancer transcriptomes. PLoS ONE 6, e17305 (2011).
Ozsolak, F. et al. Amplification-free digital gene expression profiling from minute cell quantities. Nature Methods 7, 619–621 (2010).
Chen, S. et al. De novo analysis of transcriptome dynamics in the migratory locust during the development of phase traits. PLoS ONE 5, e15633 (2010).
Schwartz, T. S. et al. A garter snake transcriptome: pyrosequencing, de novo assembly, and sex-specific differences. BMC Genomics 11, 694 (2010).
Passalacqua, K. D. et al. Structure and complexity of a bacterial transcriptome. J. Bacteriol. 191, 3203–3211 (2009).
Dalloul, R. A. et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 8, e1000475 (2010).
Jackman, S. D. & Birol, I. Assembling genomes using short-read sequencing technology. Genome Biol. 11, 202 (2010).
Rodrigue, S. et al. Unlocking short read sequencing for metagenomics. PLoS ONE 5, e11840 (2010).
Shi, H., Schmidt, B., Liu, W. & Muller-Wittig, W. A parallel algorithm for error correction in high-throughput short-read data on CUDA-enabled graphics hardware. J. Comput. Biol. 17, 603–615 (2010).
Kelley, D. R., Schatz, M. C. & Salzberg, S. L. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 11, R116 (2010).
Falgueras, J. et al. SeqTrim: a high-throughput pipeline for pre-processing any type of sequence read. BMC Bioinformatics 11, 38 (2010).
Lassmann, T., Hayashizaki, Y. & Daub, C. O. TagDust—a program to eliminate artifacts from next generation sequencing data. Bioinformatics 25, 2839–2840 (2009).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009).
Au, K. F., Jiang, H., Lin, L., Xing, Y. & Wong, W. H. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 38, 4570–4578 (2010).
Wang, K. et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 38, e178 (2010).
Wu, T. D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26, 873–881 (2010).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nature Methods 5, 621–628 (2008).
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008).
Perkins, T. T. et al. A strand-specific RNA-seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 5, e1000569 (2009).
Ozsolak, F. et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143, 1018–1029 (2010).
Salzberg, S. L. & Yorke, J. A. Beware of mis-assembled genomes. Bioinformatics 21, 4320–4321 (2005). This study highlights the importance of having standardized metrics to assess the quality of NGS assemblies.
Kinsella, M., Harismendy, O., Nakano, M., Frazer, K. A. & Bafna, V. Sensitive gene fusion detection using ambiguously mapping RNA-seq read pairs. Bioinformatics 27, 1068–1075 (2011).
McPherson, A. et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-seq data. PLoS Comput. Biol. 7, e1001138 (2011).
Tomlins, S. A. et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448, 595–599 (2007).
Pevzner, P. A., Tang, H. & Waterman, M. S. An Eulerian path approach to DNA fragment assembly. Proc. Natl Acad. Sci. USA 98, 9748–9753 (2001). This paper introduces the idea of using a De Bruijn graph for the purposes of assembly.
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nature Biotech. 29, 644–652 (2011). The Trinity de novo assembly program is introduced in this paper. This was the first NGS transcriptome assembly strategy not to rely on a genome assembler while also addressing the assembly of alternative isoforms.
Burset, M., Seledtsov, I. A. & Solovyev, V. V. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 28, 4364–4375 (2000).
Jager, M. et al. Composite transcriptome assembly of RNA-seq data in a sheep model for delayed bone healing. BMC Genomics 12, 158 (2011).
Cocquet, J., Chong, A., Zhang, G. & Veitia, R. A. Reverse transcriptase template switching and false alternative transcripts. Genomics 88, 127–131 (2006).
Haas, B. J. & Zody, M. C. Advancing RNA-seq analysis. Nature Biotech. 28, 421–423 (2010).
Greninger, A. L. et al. A metagenomic analysis of pandemic influenza A (2009 H1N1) infection in patients from North America. PLoS ONE 5, e13381 (2010).
Mizuno, H. et al. Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genomics 11, 683 (2010).
Twine, N. A., Janitz, K., Wilkins, M. R. & Janitz, M. Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer's disease. PLoS ONE 6, e16266 (2011).
Meader, S., Hillier, L. W., Locke, D., Ponting, C. P. & Lunter, G. Genome assembly quality: assessment and improvement using the neutral indel model. Genome Res. 20, 675–84 (2010).
Schaefer, B. C. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal. Biochem. 227, 255–273 (1995).
Taylor, R. C. An overview of the Hadoop/MapReduce/HBase framework and its current applications in bioinformatics. BMC Bioinformatics 11 (Suppl. 12), S1 (2010).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Acknowledgements
The work conducted by the US Department of Energy (DOE) Joint Genome Institute is supported by the Office of Science of the DOE under contract number DE-AC02-05CH11231. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the United States government, or any agency thereof, or the Regents of the University of California.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- RNA sequencing
-
(RNA-seq). An experimental protocol that uses next-generation sequencing technologies to sequence the RNA molecules within a biological sample in an effort to determine the primary sequence and relative abundance of each RNA.
- Sequencing depth
-
The average number of reads representing a given nucleotide in the reconstructed sequence. A 10× sequence depth means that each nucleotide of the transcript was sequenced, on average, ten times.
- Paired-end protocol
-
A library construction and sequencing strategy in which both ends of a DNA fragment are sequenced to produce pairs of reads (mate pairs).
- Contigs
-
An abbreviation for contiguous sequences that is used to indicate a contiguous piece of DNA assembled from shorter overlapping sequence reads.
- Low-complexity reads
-
Short DNA sequences composed of stretches of homopolymer nucleotides or simple sequence repeats.
- Quality scores
-
An integer representing the probability that a given base in a nucleic acid sequence is correct.
- k-mer frequency
-
The number of times that each k-mer (that is, a short oligonucleotide of length k) appears in a set of DNA sequences.
- Splice-aware aligner
-
A program that is designed to align cDNA reads to a genome.
- Traversing
-
A method for systematically visiting all nodes in a mathematical graph.
- Seed-and-extend aligners
-
An alignment strategy that first builds a hash table containing the location of each k-mer (seed) within the reference genome. These algorithms then extend these seeds in both directions to find the best alignment (or alignments) for each read.
- Burrows–Wheeler transform
-
(BWT). This reorders the characters within a sequence, which allows for better data compression. Many short-read aligners implement this transform in order to use less memory when aligning reads to a genome.
- Parallel computing
-
A computer programming model for distributing data processing across multiple processors, so that multiple tasks can be carried out simultaneously.
- Trans-spliced genes
-
Genes whose transcripts are created by the splicing together of two precursor mRNAs to form a single mature mRNA.
- De Bruijn graph
-
A directed mathematical graph that uses a sequence of letters of length k to represent nodes. Pairs of nodes are connected if shifting a sequence by one character creates an exact k–1 overlap between the two sequences.
- Greedily assembling
-
The use of an algorithm that joins overlapping reads together by making a series of locally optimal solutions. This strategy usually leads to a globally suboptimal solution.
- N50 size
-
The size at which half of all assembled bases reside in contigs of this size or longer.
- RACE
-
An experimental protocol termed Rapid Amplification of cDNA Ends, which is used to determine the start and end points of gene transcription.
- Cloud computing
-
The abstraction of underlying hardware architectures (for example, servers, storage and networking) to a shared pool of computing resources that can be readily provisioned and released.
Rights and permissions
About this article
Cite this article
Martin, J., Wang, Z. Next-generation transcriptome assembly. Nat Rev Genet 12, 671–682 (2011). https://doi.org/10.1038/nrg3068
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3068
This article is cited by
-
New biomarkers underlying acetic acid tolerance in the probiotic yeast Saccharomyces cerevisiae var. boulardii
Applied Microbiology and Biotechnology (2024)
-
Physiological, metabolomic, and transcriptomic reveal metabolic pathway alterations in Gymnocypris przewalskii due to cold exposure
BMC Genomics (2023)
-
Differential transcriptome response following infection of porcine ileal enteroids with species A and C rotaviruses
Virology Journal (2023)
-
Dysregulation of hypoxia-inducible factor 1α in the sympathetic nervous system accelerates diabetic cardiomyopathy
Cardiovascular Diabetology (2023)
-
Regenerative capacity of trophoblast stem cell-derived extracellular vesicles on mesenchymal stem cells
Biomaterials Research (2023)