Key Points
-
Chromatin mobility in eukaryotic nuclei allows rapid localization of genes in response to physiological stimuli.
-
Heritably repressed chromatin is found to be stably associated with non-pore elements of the nuclear envelope in organisms ranging from yeast to humans.
-
A subset of stress-induced genes in yeast have been shown to associate with nuclear pores during and after induction.
-
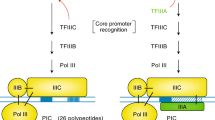
Pore-association mechanisms vary among genes, but involve both promoter elements and mRNA-surveillance and -export factors.
-
Nuclear-pore association can entail a two- to threefold increase in expression efficiency for some genes, and a more rapid reinduction after shut-off for others.
-
In male flies, gene expression on the X chromosome is enhanced twofold by the male-specific lethal (MSL) complex. This complex is associated with MTOR (Megator), a pore-associated factor.
-
MTOR and the yeast homologue MLP1 (myosin-like protein 1) are involved in mRNA elongation, which might therefore be regulated by nuclear pores.
Abstract
Cells have evolved sophisticated multi-protein complexes that can regulate gene activity at various steps of the transcription process. Recent advances highlight the role of nuclear positioning in the control of gene expression and have put nuclear envelope components at centre stage. On the inner face of the nuclear envelope, active genes localize to nuclear-pore structures whereas silent chromatin localizes to non-pore sites. Nuclear-pore components seem to not only recruit the RNA-processing and RNA-export machinery, but contribute a level of regulation that might enhance gene expression in a heritable manner.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Taddei, A., Hediger, F., Neumann, F. R. & Gasser, S. M. The function of nuclear architecture: a genetic approach. Annu. Rev. Genet, 38, 305–345 (2004).
Felsenfeld, G. & Groudine, M. Controlling the double helix. Nature 421, 448–453 (2003).
Grewal, S. I. & Elgin, S. C. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12, 178–187 (2002).
Gilbert, N., Gilchrist, S. & Bickmore, W. A. Chromatin organization in the mammalian nucleus. Int. Rev. Cytol. 242, 283–336 (2005).
Fisher, A. G. & Merkenschlager, M. Gene silencing, cell fate and nuclear organisation. Curr. Opin. Genet. Dev. 12, 193–197 (2002).
Spector, D. L. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72, 573–608 (2003).
Kosak, S. T. & Groudine, M. Form follows function: the genomic organization of cellular differentiation. Genes Dev. 18, 1371–1384 (2004).
Burgess-Beusse, B. et al. The insulation of genes from external enhancers and silencing chromatin. Proc. Natl Acad. Sci. USA 99, S16433–S16437 (2002).
Ishii, K., Arib, G., Lin, C., Van Houwe, G. & Laemmli, U. K. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109, 551–562 (2002).
Noma, K., Cam, H. P., Maraia, R. J. & Grewal, S. I. A role for TFIIIC transcription factor complex in genome organization. Cell 125, 859–872 (2006).
Gerasimova, T. I., Byrd, K. & Corces, V. G. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6, 1025–1035 (2000).
Bantignies, F., Grimaud, C., Lavrov, S., Gabut, M. & Cavalli, G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 17, 2406–2420 (2003).
Xu, Q., Li, M., Adams, J. & Cai, H. N. Nuclear location of a chromatin insulator in Drosophila melanogaster. J. Cell Sci. 117, 1025–1032 (2004).
Yusufzai, T. M., Tagami, H., Nakatani, Y. & Felsenfeld, G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13, 291–298 (2004).
Donze, D. & Kamakaka, R. T. RNA polymerase III and RNA pol II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20, 520–531 (2001).
Schmid, M. et al. Nup–PI: the nucleopore–promoter interaction of genes in yeast. Mol. Cell 21, 379–391 (2006). This paper uses a technique involving the fusion of a pore protein to MN to show that promoters associate with nuclear pores in yeast.
Casolari, J. M. et al. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117, 427–439 (2004). The initial cross-linking study, showing that a range of pore proteins can be crosslinked by formaldehyde to many genes, among which are those induced by galactose.
Taddei, A., Hediger, F., Neumann, F. R., Bauer, C. & Gasser, S. M. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23, 1301–1312 (2004). Sir4, yKu and Esc1 are able to relocate DNA to the nuclear envelope independently of their silencing function. The authors descibe interdependencies between proteins. Esc1 is found to be co-localized with silent chromatin but distinct from nuclear pores.
Gartenberg, M. R. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr. Opin. Microbiol. 3, 132–137 (2000).
Palladino, F. et al. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75, 543–555 (1993).
Gotta, M. et al. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134, 1349–1363 (1996).
Andrulis, E. D. et al. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 22, 8292–8301 (2002).
Gartenberg, M. R., Neumann, F. R., Laroche, T., Blaszczyk, M. & Gasser, S. M. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119, 955–967 (2004). Active and inactive chromatin moves freely through the nucleoplasm once it is released from the chromosome. Sir-mediated silencing is sufficient to anchor DNA at the nuclear envelope, and is not necessary if Sir proteins are dispersed.
Hediger, F., Neumann, F. R., Van Houwe, G., Dubrana, K. & Gasser, S. M. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12, 2076–2089 (2002).
Hediger, F., Dubrana, K. & Gasser, S. M. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J. Struct. Biol. 140, 79–91 (2002).
Zhao, X. & Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad. Sci. USA 102, 4777–4782 (2005).
Zhao, X., Wu, C. Y. & Blobel, G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167, 605–611 (2004).
Hiraga, S., Robertson, E. D. & Donaldson, A. D. The Ctf18 RFC-like complex positions yeast telomeres but does not specify their replication time. EMBO J. 25, 1505–1514 (2006).
Maillet, L. et al. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10, 1796–1811 (1996). Telomere foci favour the repression of silencer-bound constructs by sequestering Sir proteins, which are shown to be limiting for repression.
Andrulis, E. D., Neiman, A. M., Zappulla, D. C. & Sternglanz, R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394, 592–595 (1998). Anchoring of a gene to the nuclear envelope can enhance silencer function in yeast.
Capell, B. C. & Collins, F. S. Human laminopathies: nuclei gone genetically awry. Nature Rev. Genet. 7, 940–952 (2006).
Sullivan, T. et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913–920 (1999).
Shumaker, D. K. et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl Acad. Sci. USA 103, 8703–8708 (2006).
Scaffidi, P. & Misteli, T. Reversal of the cellular phenotype in the premature aging disease Hutchinson–Gilford progeria syndrome. Nature Med. 11, 440–445 (2005).
Goldman, R. D. et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc. Natl Acad. Sci. USA 101, 8963–8968 (2004).
Liu, J. et al. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 100, 4598–4603 (2003).
Pickersgill, H. et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nature Genet. 38, 1005–1014 (2006). This work describes the binding profile of D. melanogaster lamins, showing that lamins preferentially bind to transcriptionally inactive genes.
Paddy, M. R., Belmont, A. S., Saumweber, H., Agard, D. A. & Sedat, J. W. Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell 62, 89–106 (1990).
Marshall, W. F. et al. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7, 930–939 (1997). The first description of the constrained random walk of LacO-tagged yeast and D. melanogaster loci. This work shows that a centromere-proximal locus is constrained in yeast.
Heun, P., Laroche, T., Shimada, K., Furrer, P. & Gasser, S. M. Chromosome dynamics in the yeast interphase nucleus. Science 294, 2181–2186 (2001). Silent telomeres and active non-telomeric loci are shown to have different amounts of spatial constraint in their random-walk movement in the yeast nucleus. This work coupled Nup49–GFP with LacO-tagged loci to measure the absolute movement of genes.
Bystricky, K., Heun, P., Gehlen, L., Langowski, J. & Gasser, S. M. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl Acad. Sci. USA 101, 16495–16500 (2004).
Heun, P., Laroche, T., Raghuraman, M. K. & Gasser, S. M. The positioning and dynamics of origins of replication in the budding yeast nucleus. J. Cell Biol. 152, 385–400 (2001).
Jin, Q., Trelles-Sticken, E., Scherthan, H. & Loidl, J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141, 21–29 (1998).
Sage, D., Neumann, F. R., Hediger, F., Gasser, S. M. & Unser, M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans. Image Process. 14, 1372–1383 (2005).
Rosa, A., Maddocks, J. H., Neumann, F. R., Gasser, S. M. & Stasiak, A. Measuring limits of telomere movement on nuclear envelope. Biophys. J. 90, L24–L26 (2006).
Taddei, A. et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441, 774–778 (2006). Telomeric foci and pores are distinct functional compartments. A subtelomeric gene shifts from a telomeric focus to a pore upon induction on low glucose. When this relocation is impaired, maximal induction is prevented.
Cabal, G. G. et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441, 770–773 (2006). Stimulation of galactose-inducible promoters correlates with relocalization to the nuclear periphery, which is mediated by components of the SAGA complex in yeast.
Bystricky, K., Laroche, T., van Houwe, G., Blaszczyk, M. & Gasser, S. M. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J. Cell Biol. 168, 375–387 (2005).
Marshall, W. F., Fung, J. C. & Sedat, J. W. Deconstructing the nucleus: global architecture from local interactions. Curr. Opin. Genet. Dev. 7, 259–263 (1997).
Chubb, J. R., Boyle, S., Perry, P. & Bickmore, W. A. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439–445 (2002).
Janicki, S. M. et al. From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 (2004). The first live tracking of an mRNA from a gene to the nuclear pore. The process is rapid and occurs with a random-walk character.
Misteli, T. The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181–185 (2001).
Menon, B. B. et al. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl Acad. Sci. USA 102, 5749–5754 (2005).
Casolari, J. M., Brown, C. R., Drubin, D. A., Rando, O. J. & Silver, P. A. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 19, 1188–1198 (2005).
Brickner, D. G. et al. H2A. Z-mediated Localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5, e81 (2007). Active genes are retained at the nuclear periphery for several generations. The retention and reactivation requires histone variant H2A.Z. The authors propose that such a mechanism could provide a system by which cells remember the status of the actively transcribed gene through cell generations.
Brickner, J. H. & Walter, P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2, e342 (2004). Ino1 expression requires contact with the nuclear envelope for promoter activation.
Dieppois, G., Iglesias, N. & Stutz, F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol. Cell. Biol. 26, 7858–7870 (2006).
Abruzzi, K. C., Belostotsky, D. A., Chekanova, J. A., Dower, K. & Rosbash, M. 3′-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation. EMBO J. 25, 4253–4262 (2006). 3′ UTR sequences and mRNA influence prolonged gene positioning at nuclear pores.
Rodriguez-Navarro, S. et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116, 75–86 (2004).
Fischer, T. et al. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21, 5843–5852 (2002).
Strasser, K. et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308 (2002).
O'Sullivan, J. M. et al. Gene loops juxtapose promoters and terminators in yeast. Nature Genet. 36, 1014–1018 (2004).
Ansari, A. & Hampsey, M. A role for the CPF 3′-end processing machinery in RNAPII-dependent gene looping. Genes Dev. 19, 2969–2978 (2005).
Furger, A., O'Sullivan, J. M., Binnie, A., Lee, B. A. & Proudfoot, N. J. Promoter proximal splice sites enhance transcription. Genes Dev. 16, 2792–2799 (2002).
Proudfoot, N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16, 272–278 (2004).
Mendjan, S. et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21, 811–823 (2006). The first report of the novel biochemical and functional link of MTOR and NUP153 to dosage compensation of the male X chromosome in D. melanogaster .
Jin, Y., Wang, Y., Johansen, J. & Johansen, K. M. JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associated with the male specific lethal (MSL) dosage compensation complex. J. Cell Biol. 149, 1005–1010 (2000).
Deuring, R. et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5, 355–365 (2000).
Corona, D. F., Clapier, C. R., Becker, P. B. & Tamkun, J. W. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 3, 242–247 (2002).
Furuhashi, H., Nakajima, M. & Hirose, S. DNA supercoiling factor contributes to dosage compensation in Drosophila. Development 133, 4475–4483 (2006).
Lucchesi, J. C., Kelly, W. G. & Panning, B. Chromatin remodeling in dosage compensation. Annu Rev Genet 39, 615–651 (2005).
Mendjan, S. & Akhtar, A. The right dose for every sex. Chromosoma 116, 95–106 (2006).
Straub, T. & Becker, P. B. Dosage compensation: the beginning and end of generalization. Nature Rev. Genet. 8, 47–57 (2007).
Alekseyenko, A. A., Larschan, E., Lai, W. R., Park, P. J. & Kuroda, M. I. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 20, 848–857 (2006).
Gilfillan, G. D. et al. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 20, 858–870 (2006).
Legube, G., McWeeney, S. K., Lercher, M. J. & Akhtar, A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 20, 871–883 (2006).
Smith, E. R., Allis, C. D. & Lucchesi, J. C. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 276, 31483–31486 (2001). References 74–77 use a ChIP–chip strategy to identify the binding profile of MSL proteins, and show that the MSL complex is enriched on X-linked genes with preferential binding towards the 3′ end of genes.
Hamada, F. N., Park, P. J., Gordadze, P. R. & Kuroda, M. I. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 19, 2289–2294 (2005).
Straub, T., Gilfillan, G. D., Maier, V. K. & Becker, P. B. The Drosophila MSL complex activates the transcription of target genes. Genes Dev. 19, 2284–2288 (2005). References 78 and 79 collectively show that transcription of MSL-bound X-linked genes is affected by depletion of MSL2 in D. melanogaster cells.
Galy, V. et al. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63–73 (2004).
Sommer, P. & Nehrbass, U. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr. Opin. Cell Biol. 17, 294–301 (2005).
Libri, D. et al. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell Biol. 22, 8254–8266 (2002).
Zenklusen, D., Vinciguerra, P., Wyss, J. C. & Stutz, F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22, 8241–8253 (2002).
Andrulis, E. D. et al. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420, 837–841 (2002). This work describes the co-purification of the RNA pol II complex with components of the nuclear exosome, thus linking the RNA-processing machinery with the transcription machinery.
Hieronymus, H., Yu, M. C. & Silver, P. A. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 18, 2652–2662 (2004).
Hilleren, P., McCarthy, T., Rosbash, M., Parker, R. & Jensen, T. H. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413, 538–542 (2001).
Herrera, F. J. & Triezenberg, S. J. Molecular biology: what ubiquitin can do for transcription. Curr. Biol. 14, R622–R624 (2004).
Vazquez, J., Belmont, A. S. & Sedat, J. W. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12, 1473–1483 (2002).
Cremer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature Rev. Genet. 2, 292–301 (2001).
Lanctot, C., Cheutin, T., Cremer, M., Cavalli, G. & Cremer, T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature Rev. Genet. 8, 104–115 (2007).
Chambeyron, S. & Bickmore, W. A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18, 1119–1130 (2004).
Chambeyron, S., Da Silva, N. R., Lawson, K. A. & Bickmore, W. A. Nuclear re-organisation of the HOXB complex during mouse embryonic development. Development 132, 2215–2223 (2005).
Morey, C., Da Silva, N. R., Perry, P. & Bickmore, W. A. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development 134, 909–919 (2007).
Tran, E. J. & Wente, S. R. Dynamic nuclear pore complexes: life on the edge. Cell 125, 1041–1053 (2006).
Vinciguerra, P., Iglesias, N., Camblong, J., Zenklusen, D. & Stutz, F. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 24, 813–823 (2005).
Acknowledgements
We thank members of our laboratories for support. We are very grateful to Jop Kind for Figure 5. We apologize to any colleagues whose work could not be cited owing to space limitations. This work was support by EU funding to A.A. and S.M.G. S.M.G. acknowledges support from the Novartis Research Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Nuclear periphery
-
A term that generally refers to the nuclear-membrane bilayer, its associated proteins and the embedded nuclear-pore complexes.
- Dosage compensation
-
A phenomenon that ensures equalized gene expression of X-chromosomal genes between males and females. In Drosophila melanogaster, this results in approximately twofold higher levels of transcriptional activation in the single male X chromosome compared with the female X chromosomes.
- Nuclear lamina
-
A meshwork of a nuclear intermediate filament protein that is found at the interface between the inner nuclear membrane and chromatin.
- Fluorescence in situ hybridization
-
A technique whereby a fluorescently labelled DNA probe is used to detect a particular chromosomal region by fluorescence microscopy.
- Chromatin immunoprecipitation
-
A technique that involves crosslinking methods and is used to identify pieces of DNA or chromatin that contact a protein of interest in vivo.
- Nuclear-pore complexes
-
Large, multiprotein complexes (composed of about 30 proteins) that are embedded in the nuclear membrane and serve as gateways for traffic between the nucleus and the cytoplasm.
- MSL complex
-
An RNA–protein complex containing at least five male-specific Lethal (MSL) proteins, MSL1, MSL2, MSL3, MOF and MLE, and two non-coding RNAs, roX1 and roX2. This complex is stably expressed in male flies and regulates dosage compensation of X-linked genes.
Rights and permissions
About this article
Cite this article
Akhtar, A., Gasser, S. The nuclear envelope and transcriptional control. Nat Rev Genet 8, 507–517 (2007). https://doi.org/10.1038/nrg2122
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg2122
This article is cited by
-
Polymeric nature of tandemly repeated genes enhances assembly of constitutive heterochromatin in fission yeast
Communications Biology (2023)
-
The inner nuclear membrane protein Lem2 coordinates RNA degradation at the nuclear periphery
Nature Structural & Molecular Biology (2022)
-
SETDB1-like MET-2 promotes transcriptional silencing and development independently of its H3K9me-associated catalytic activity
Nature Structural & Molecular Biology (2022)
-
Functional mechanisms and abnormalities of the nuclear lamina
Nature Cell Biology (2021)
-
Nup133 and ERα mediate the differential effects of hyperoxia-induced damage in male and female OPCs
Molecular and Cellular Pediatrics (2020)