Key Points
-
The human genome is pervasively transcribed. Most RNA transcripts represent non-coding RNAs (ncRNAs), of which there are many categories. This article focuses on long non-coding RNAs (lncRNAs).
-
Many lncRNAs have been shown to be functional and/or related to several human diseases, including various forms of cancer and inheritable diseases such as Huntington's disease and Fragile X syndrome. lncRNAs can positively or negatively regulate gene expression and chromatin architecture.
-
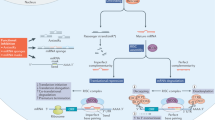
Natural antisense transcripts (NATs) are highly abundant in the human genome and found in many known gene loci where they are transcribed in the opposite direction of conventional protein-coding genes.
-
NATs frequently regulate the expression of protein-coding genes, either positively or negatively. Most NATs mediate transcriptional repression at the chromatin level.
-
AntagoNATs are oligonucleotides that specifically target NATs and are capable of upregulating the expression of corresponding protein coding genes in vitro as well as in vivo.
-
AntagoNATs induce gene upregulation in a gene-locus-specific and reversible manner.
-
AntagoNAT upregulation technology has been applied to the upregulation of genes including growth factors, tumour suppressors, transcription factors and those genes that are deficient in genetic diseases.
-
There is a scarcity of other methods for inducing locus-specific and reversible gene upregulation.
Abstract
The majority of currently available drugs and tool compounds exhibit an inhibitory mechanism of action and there is a relative lack of pharmaceutical agents that are capable of increasing the activity of effectors or pathways for therapeutic benefit. Indeed, the upregulation of many genes, including tumour suppressors, growth factors, transcription factors and genes that are deficient in various genetic diseases, would be desired in specific situations. Recently, key roles for regulatory long non-coding RNAs (lncRNAs) in the regulation of gene expression have begun to emerge. lncRNAs can positively or negatively regulate gene expression and chromatin architecture. Here, we review the current understanding of the mechanisms of action of lncRNAs and their roles in disease, focusing on recent work in the design of inhibitors of the natural antisense transcript (NAT) class of lncRNAs, known as antagoNAT oligonucleotides, and the issues associated with their potential therapeutic application.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
29 November 2013
The second paragraph in the section titled "Challenges in targeting long non-coding RNAs" on page 442 of this Review was inserted during manuscript revision. The wording of this paragraph drew heavily on, and mostly repeated, a paragraph in an earlier review article by J. T. Lee (Lee, J. T. Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439; 2012). Although the Science review was cited elsewhere in this article, it should also have been cited at this point in order to attribute correctly the ideas expressed by Lee. The author apologizes for introducing this passage, and for the lack of citation to the paper from which it derived, and is pleased to correct the record accordingly.
References
Byrne, B. J., Falk, D. J., Clement, N. & Mah, C. S. Gene therapy approaches for lysosomal storage disease: next-generation treatment. Hum. Gene Ther. 23, 808–815 (2012).
Sheridan, C. Gene therapy finds its niche. Nature Biotech. 29, 121–128 (2011).
Ciesielska, A. et al. Cerebral infusion of AAV9 vector-encoding non-self proteins can elicit cell-mediated immune responses. Mol. Ther. 21, 158–166 (2013).
Unzu, C. et al. Transient and intensive pharmacological immunosuppression fails to improve AAV-based liver gene transfer in non-human primates. J. Transl. Med. 10, 122 (2012).
Waehler, R., Russell, S. J. & Curiel, D. T. Engineering targeted viral vectors for gene therapy. Nature Rev. Genet. 8, 573–587 (2007).
Mingozzi, F. & High, K. A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nature Rev. Genet. 12, 341–355 (2011).
Thrasher, A. J. et al. Gene therapy: X-SCID transgene leukaemogenicity. Nature 443, e5–e6 (2006).
Gaspar, H. B. et al. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci. Transl. Med. 3, 97ra79 (2011).
Woods, N. B., Bottero, V., Schmidt, M., von Kalle, C. & Verma, I. M. Gene therapy: therapeutic gene causing lymphoma. Nature 440, 1123 (2006).
Yla-Herttuala, S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther. 20, 1831–1832 (2012).
Overington, J. P., Al-Lazikani, B. & Hopkins, A. L. How many drug targets are there? Nature Rev. Drug Discov. 5, 993–996 (2006).
Karamouzis, M. V. & Papavassiliou, A. G. Transcription factor networks as targets for therapeutic intervention of cancer: the breast cancer paradigm. Mol. Med. 17, 1133–1136 (2011).
Libermann, T. A. & Zerbini, L. F. Targeting transcription factors for cancer gene therapy. Curr. Gene Ther. 6, 17–33 (2006).
Arrowsmith, C. H., Bountra, C., Fish, P. V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nature Rev. Drug Discov. 11, 384–400 (2012).
Dawson, M. A. & Kouzarides, T. Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27 (2012).
Lindow, M. & Kauppinen, S. Discovering the first microRNA-targeted drug. J. Cell Biol. 199, 407–412 (2012).
Thum, T. MicroRNA therapeutics in cardiovascular medicine. EMBO Mol. Med. 4, 3–14 (2012).
Broderick, J. A. & Zamore, P. D. MicroRNA therapeutics. Gene Ther. 18, 1104–1110 (2011).
Wang, V. & Wu, W. MicroRNA-based therapeutics for cancer. BioDrugs 23, 15–23 (2009).
Sun, W., Julie Li, Y. S., Huang, H. D., Shyy, J. Y. & Chien, S. microRNA: a master regulator of cellular processes for bioengineering systems. Annu. Rev. Biomed. Eng. 12, 1–27 (2010).
Wahlestedt, C. Natural antisense and noncoding RNA transcripts as potential drug targets. Drug Discov. Today 11, 503–508 (2006). This is the first non-patent publication to describe the antagoNAT strategy.
Schwartz, J. C. et al. Antisense transcripts are targets for activating small RNAs. Nature Struct. Mol. Biol. 15, 842–848 (2008).
Morris, K. V. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics 4, 296–301 (2009).
Carninci, P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005). This study describes the large-scale effort required to produce data showing that the mammalian genome is pervasively transcribed.
Cheng, J. et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154 (2005).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012). This study also describes the large-scale effort required to produce data showing that the mammalian genome is pervasively transcribed.
Derrien, T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012).
Mattick, J. S. The functional genomics of noncoding RNA. Science 309, 1527–1528 (2005).
Graifer, D. & Karpova, G. Structural and functional topography of the human ribosome. Acta Biochim. Biophys. Sin. 44, 281–299 (2012).
St Laurent, G. et al. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genomics 13, 504 (2012).
Brown, C. J. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44 (1991).
Lee, J. T. Epigenetic regulation by long noncoding RNAs. Science 338, 1435–1439 (2012).
Taft, R. J., Pheasant, M. & Mattick, J. S. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 29, 288–299 (2007).
Mathews, D. H., Moss, W. N. & Turner, D. H. Folding and finding RNA secondary structure. Cold Spring Harb. Perspect. Biol. 2, a003665 (2010).
Wahlestedt, C. et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl Acad. Sci. USA 97, 5633–5638 (2000).
Khalil, A. M., Faghihi, M. A., Modarresi, F., Brothers, S. P. & Wahlestedt, C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS ONE 3, e1486 (2008).
Katayama, S. et al. Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566 (2005). This paper describes a large-scale effort to show that extensive and functionally relevant antisense transcription occurs in the mammalian genome.
Engstrom, P. G. et al. Complex loci in human and mouse genomes. PLoS Genet. 2, e47 (2006).
Khalil, A. M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA 106, 11667–11672 (2009). This paper demonstrates that many lncRNAs are not associated with protein-coding loci.
Guttman, M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009).
Guttman, M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011). This is a large-scale demonstration showing that the function of many lncRNAs is not associated with protein-coding loci.
Kretz, M. et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235 (2013).
Nakaya, H. I. et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 8, R43 (2007).
Faghihi, M. A. et al. RNAi screen indicates widespread biological function for human natural antisense transcripts. PLoS ONE 5, e13177 (2010). This is a large-scale demonstration of the function of many long non-coding NATs.
Faghihi, M. A. & Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nature Rev. Mol. Cell Biol. 10, 637–643 (2009).
Magistri, M., Faghihi, M. A., St Laurent, G. & Wahlestedt, C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 28, 389–396 (2012).
Faghihi, M. A. & Wahlestedt, C. RNA interference is not involved in natural antisense mediated regulation of gene expression in mammals. Genome Biol. 7, R38 (2006).
Carrieri, C. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012).
Mohammad, F., Mondal, T. & Kanduri, C. Epigenetics of imprinted long noncoding RNAs. Epigenetics 4, 277–286 (2009).
Yang, L. et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788 (2011).
Rinn, J. L. et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007).
Tsai, M. C. et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010).
Hung, T. et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature Genet. 43, 621–629 (2011).
Huarte, M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010).
Feng, J. et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484 (2006).
Wang, X. et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130 (2008).
Tripathi, V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010).
Eissmann, M. et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 9, 1076–1087 (2012).
Kino, T., Hurt, D. E., Ichijo, T., Nader, N. & Chrousos, G. P. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3, ra8 (2010).
Bond, C. S. & Fox, A. H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644 (2009).
Clemson, C. M. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358 (2011).
Cesana, M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011).
Kim, T. K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010).
Sahu, B. et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 30, 3962–3976 (2011).
Wang, D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011).
Yap, K. L. et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674 (2010).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010).
Yang, Z. et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 18, 1243–1250 (2011).
Kogo, R. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326 (2011).
Niinuma, T. et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 72, 1126–1136 (2012).
Gutschner, T. & Diederichs, S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 9, 703–719 (2012).
Luo, J. H. et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology 44, 1012–1024 (2006).
Schmidt, L. H. et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol. 6, 1984–1992 (2011).
Geng, Y. J., Xie, S. L., Li, Q., Ma, J. & Wang, G. Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res. 39, 2119–2128 (2011).
Kim, K. et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32, 1616–1625 (2012).
Silva, J. M., Boczek, N. J., Berres, M. W., Ma, X. & Smith, D. I. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 8, 496–505 (2011).
Jendrzejewski, J. et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc. Natl Acad. Sci. USA 109, 8646–8651 (2012).
Han, Y., Liu, Y., Gui, Y. & Cai, Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J. Surg. Oncol. 107, 555–559 (2012).
Tsang, W. P., Wong, T. W., Cheung, A. H., Co, C. N. & Kwok, T. T. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA 13, 890–898 (2007).
Qureshi, I. A., Mattick, J. S. & Mehler, M. F. Long non-coding RNAs in nervous system function and disease. Brain Res. 1338, 20–35 (2010).
Pastori, C. & Wahlestedt, C. Involvement of long noncoding RNAs in diseases affecting the central nervous system. RNA Biol. 9, 860–870 (2012).
St Laurent, G. & Wahlestedt, C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 30, 612–621 (2007).
Mirkin, S. M. Expandable DNA repeats and human disease. Nature 447, 932–940 (2007).
Moseley, M. L. et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nature Genet. 38, 758–769 (2006).
Cho, D. H. et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell 20, 483–489 (2005).
Faghihi, M. A. et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nature Med. 14, 723–730 (2008).
Mus, E., Hof, P. R. & Tiedge, H. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc. Natl Acad. Sci. USA 104, 10679–10684 (2007).
Johnson, R. et al. The human accelerated region 1 noncoding RNA is repressed by REST in Huntington's disease. Physiol. Genomics 41, 269–274 (2010).
Troy, A. & Sharpless, N. E. Genetic “lnc”-age of noncoding RNAs to human disease. J. Clin. Invest. 122, 3837–3840 (2012).
van Dijk, M. et al. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J. Clin. Invest. 122, 4003–4011 (2012).
Maass, P. G. et al. A misplaced lncRNA causes brachydactyly in humans. J. Clin. Invest. 122, 3990–4002 (2012).
Millar, J. K. et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum. Mol. Genet. 9, 1415–1423 (2000).
Ladd, P. D. et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 16, 3174–3187 (2007).
Cabianca, D. S. et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149, 819–831 (2012).
Wapinski, O. & Chang, H. Y. Long noncoding RNAs and human disease. Trends Cell Biol. 21, 354–361 (2011).
Modarresi, F. et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nature Biotech. 30, 453–459 (2012). This is the first in vivo demonstration of the efficacy of antagoNATs.
Li, L. C. et al. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA 103, 17337–17342 (2006).
Janowski, B. A. et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nature Chem. Biol. 3, 166–173 (2007).
Scheele, C. et al. The human PINK1 locus is regulated in vivo by a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics 8, 74 (2007).
Hopkins, A. L. & Groom, C. R. The druggable genome. Nature Rev. Drug Discov. 1, 727–730 (2002).
Desnick, R. J. & Schuchman, E. H. Enzyme replacement therapy for lysosomal diseases: lessons from 20 years of experience and remaining challenges. Annu. Rev. Genom. Hum. Genet. 13, 307–335 (2012).
Dirin, M. & Winkler, J. Influence of diverse chemical modifications on the ADME characteristics and toxicology of antisense oligonucleotides. Expert Opin. Biol. Ther. 2 Mar 2013 (10.1517/14712598.2013.774366).
Jones, B. K., Levorse, J. M. & Tilghman, S. M. Igf2 imprinting does not require its own DNA methylation or H19 RNA. Genes Dev. 12, 2200–2207 (1998).
Schorderet, P. & Duboule, D. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PLoS Genet. 7, e1002071 (2011).
Nakagawa, S., Naganuma, T., Shioi, G. & Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 193, 31–39 (2011).
Zhang, B. et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123 (2012).
Bennett, C. F. & Swayze, E. E. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50, 259–293 (2010).
Senn, J. J., Burel, S. & Henry, S. P. Non-CpG-containing antisense 2′-methoxyethyl oligonucleotides activate a proinflammatory response independent of Toll-like receptor 9 or myeloid differentiation factor 88. J. Pharmacol. Exp. Ther. 314, 972–979 (2005).
Krieg, A. M. Therapeutic potential of Toll-like receptor 9 activation. Nature Rev. Drug Discov. 5, 471–484 (2006).
Freier, S. & Watt, A. T. in Antisense Drug Technology: Principles, Strategies, and Applications (ed. Crooke, S. T.) 118–138 (CRC Press, 2007).
Xu, Z., Almudevar, A. & Mathews, D. H. Statistical evaluation of improvement in RNA secondary structure prediction. Nucleic Acids Res. 40, e26 (2012).
Lima, W. F. et al. Human RNase H1 discriminates between subtle variations in the structure of the heteroduplex substrate. Mol. Pharmacol. 71, 83–91 (2007).
Carroll, J. B. et al. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol. Ther. 19, 2178–2185 (2011).
Stein, C. A. The experimental use of antisense oligonucleotides: a guide for the perplexed. J. Clin. Invest. 108, 641–644 (2001).
Shim, M. S. & Kwon, Y. J. Efficient and targeted delivery of siRNA in vivo. FEBS J. 277, 4814–4827 (2010).
Huang, L. & Liu, Y. In vivo delivery of RNAi with lipid-based nanoparticles. Annu. Rev. Biomed. Eng. 13, 507–530 (2011).
Pollack, A. F.D.A. approves genetic drug to treat rare disease. New York Times [online], (2013).
Juliano, R. L., Carver, K., Cao, C. & Ming, X. Receptors, endocytosis, and trafficking: the biological basis of targeted delivery of antisense and siRNA oligonucleotides. J. Drug Target 21, 27–43 (2013).
Wahlestedt, C. et al. Antisense oligodeoxynucleotides to NMDA-R1 receptor channel protect cortical neurons from excitotoxicity and reduce focal ischaemic infarctions. Nature 363, 260–263 (1993).
Wahlestedt, C., Pich, E. M., Koob, G. F., Yee, F. & Heilig, M. Modulation of anxiety and neuropeptide Y-Y1 receptors by antisense oligodeoxynucleotides. Science 259, 528–531 (1993).
Standifer, K. M., Chien, C. C., Wahlestedt, C., Brown, G. P. & Pasternak, G. W. Selective loss of δ opioid analgesia and binding by antisense oligodeoxynucleotides to a δ opioid receptor. Neuron 12, 805–810 (1994).
Yee, F., Ericson, H., Reis, D. J. & Wahlestedt, C. Cellular uptake of intracerebroventricularly administered biotin- or digoxigenin-labeled antisense oligodeoxynucleotides in the rat. Cell. Mol. Neurobiol. 14, 475–486 (1994).
Southwell, A. L., Skotte, N. H., Bennett, C. F. & Hayden, M. R. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol. Med. 18, 634–643 (2012).
Hayek, S. M., Deer, T. R., Pope, J. E., Panchal, S. J. & Patel, V. B. Intrathecal therapy for cancer and non-cancer pain. Pain Physician 14, 219–248 (2011).
Rigo, F., Hua, Y., Krainer, A. R. & Bennett, C. F. Antisense-based therapy for the treatment of spinal muscular atrophy. J. Cell Biol. 199, 21–25 (2012).
Gommans, W. M., Haisma, H. J. & Rots, M. G. Engineering zinc finger protein transcription factors: the therapeutic relevance of switching endogenous gene expression on or off at command. J. Mol. Biol. 354, 507–519 (2005).
Klug, A. Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 293, 215–218 (1999).
Matera, A. G., Terns, R. M. & Terns, M. P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nature Rev. Mol. Cell Biol. 8, 209–220 (2007).
Bachellerie, J. P., Cavaille, J. & Huttenhofer, A. The expanding snoRNA world. Biochimie 84, 775–790 (2002).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Fabian, M. R. & Sonenberg, N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature Struct. Mol. Biol. 19, 586–593 (2012).
Rother, S. & Meister, G. Small RNAs derived from longer non-coding RNAs. Biochimie 93, 1905–1915 (2011).
Pruijn, G. J., Wingens, P. A., Peters, S. L., Thijssen, J. P. & van Venrooij, W. J. Ro RNP associated Y RNAs are highly conserved among mammals. Biochim. Biophys. Acta 1216, 395–401 (1993).
Jeon, Y. & Lee, J. T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133 (2011).
Schmitz, K. M., Mayer, C., Postepska, A. & Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269 (2010).
Martianov, I., Ramadass, A., Serra Barros, A., Chow, N. & Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445, 666–670 (2007).
Beerli, R. R. & Barbas, C. F. Engineering polydactyl zinc-finger transcription factors. Nature Biotech. 20, 135–141 (2002).
Cayre, A., Rossignol, F., Clottes, E. & Penault-Llorca, F. aHIF but not HIF-1α transcript is a poor prognostic marker in human breast cancer. Breast Cancer Res. 5, R223–230 (2003).
Thrash-Bingham, C. A. & Tartof, K. D. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J. Natl Cancer Inst. 91, 143–151 (1999).
Zolk, O., Solbach, T. F., Eschenhagen, T., Weidemann, A. & Fromm, M. F. Activation of negative regulators of the hypoxia-inducible factor (HIF) pathway in human end-stage heart failure. Biochem. Biophys. Res. Commun. 376, 315–320 (2008).
Pasmant, E. et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 67, 3963–3969 (2007).
Pasmant, E. et al. Role of noncoding RNA ANRIL in genesis of plexiform neurofibromas in neurofibromatosis type 1. J. Natl Cancer Inst. 103, 1713–1722 (2011).
Pasmant, E., Sabbagh, A., Vidaud, M. & Bieche, I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 25, 444–448 (2011).
Annilo, T., Kepp, K. & Laan, M. Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol. Biol. 10, 81 (2009).
Parenti, R., Paratore, S., Torrisi, A. & Cavallaro, S. A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during β-amyloid-induced apoptosis. Eur. J. Neurosci. 26, 2444–2457 (2007).
Zhang, H., Gao, S. & De Geyter, C. A natural antisense transcript, BOKAS, regulates the pro-apoptotic activity of human Bok. Int. J. Oncol. 34, 1135–1138 (2009).
Chung, D. W., Rudnicki, D. D., Yu, L. & Margolis, R. L. A natural antisense transcript at the Huntington's disease repeat locus regulates HTT expression. Hum. Mol. Genet. 20, 3467–3477 (2011).
Yu, W. et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206 (2008).
Ozgur, E. et al. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin. Exp. Med. 10 Apr 2012 (10.1007/s10238-012-0181-x).
Hu, W., Yuan, B., Flygare, J. & Lodish, H. F. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 25, 2573–2578 (2011).
Paralkar, V. R. & Weiss, M. J. A new 'Linc' between noncoding RNAs and blood development. Genes Dev. 25, 2555–2558 (2011).
Mourtada-Maarabouni, M., Pickard, M. R., Hedge, V. L., Farzaneh, F. & Williams, G. T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 28, 195–208 (2009).
Acknowledgements
The author apologizes to colleagues whose work could not be discussed or cited owing to space limitations. He also thanks many members of his laboratory, in particular V. Peschansky and P. Halley, for their critical review of the literature and for providing important help with the manuscript. M. Magistri and Z. Zeier have provided scientific input and expert help in designing the figures. M. A. Faghihi, P. Kapranov and G. St Laurent are acknowledged for their critical reading and/or for providing important input on RNA classification. Finally, the author thanks J. Hsiao, O. Khorkova, C. Coito, P. Frost and others at OPKO-CURNA for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Claes Wahlestedt is a consultant for Opko Health.
Related links
Glossary
- MicroRNAs
-
(miRNAs). Small non-coding RNAs that affect the stability of many mRNAs.
- Long non-coding RNAs
-
(lncRNAs). RNA transcripts greater than 200 nucleotides in length that lack an open reading frame and therefore do not encode protein.
- Natural antisense transcript
-
(NAT). An RNA transcript originating from the opposite strand of a sense (often protein-coding) RNA transcript.
- INK4B–ARF–INK4A locus
-
A locus that encodes three tumour suppressor proteins: INK4B, which is encoded by cyclin-dependent kinase inhibitor 2B (CDKN2B); and INK4A and ARF, which are encoded by CDKN2A.
- AntagoNATs
-
Oligonucleotide inhibitors targeted to a natural antisense transcript (NAT).
Rights and permissions
About this article
Cite this article
Wahlestedt, C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov 12, 433–446 (2013). https://doi.org/10.1038/nrd4018
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrd4018
This article is cited by
-
Transcriptome analysis of the cerebral cortex of acrylamide-exposed wild-type and IL-1β-knockout mice
Archives of Toxicology (2024)
-
Amplifying gene expression with RNA-targeted therapeutics
Nature Reviews Drug Discovery (2023)
-
LncRNA BCAN-AS1 stabilizes c-Myc via N6-methyladenosine-mediated binding with SNIP1 to promote pancreatic cancer
Cell Death & Differentiation (2023)
-
The Promising Epigenetic Regulators for Refractory Epilepsy: An Adventurous Road Ahead
NeuroMolecular Medicine (2023)
-
M7G-Related lncRNAs predict prognosis and regulate the immune microenvironment in lung squamous cell carcinoma
BMC Cancer (2022)