Abstract
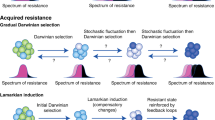
Epigenetic events, which are somatically inherited through cell division, are potential drivers of acquired drug resistance in cancer. The high rate of epigenetic change in tumours generates diversity in gene expression patterns that can rapidly evolve through drug selection during treatment, leading to the development of acquired resistance. This will potentially confound stratified chemotherapy decisions that are solely based on mutation biomarkers. Poised epigenetic states in tumour cells may drive multistep epigenetic fixation of gene expression during the acquisition of drug resistance, which has implications for clinical strategies to prevent the emergence of drug resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Agarwal, R. & Kaye, S. B. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nature Rev. Cancer 3, 502–516 (2003).
Camidge, D. R., Pao, W. & Sequist, L. V. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nature Rev. Clin. Oncol. 11, 473–481 (2014).
Pujade-Lauraine, E. et al. Pegylated liposomal doxorubicin and carboplatin compared with paclitaxel and carboplatin for patients with platinum-sensitive ovarian cancer in late relapse. J. Clin. Oncol. 28, 3323–3329 (2010).
Kaplan, B., Qazi, Y. & Wellen, J. R. Strategies for the management of adverse events associated with mTOR inhibitors. Transplant. Rev. 28, 126–133 (2014).
Shen, D.-W., Pastan, I. & Gottesman, M. M. In situ hybridization analysis of acquisition and loss of the human multidrug-resistance gene. Cancer Res. 48, 4334–4339 (1988).
Zeller, C. et al. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene 31, 4567–4576 (2012).
Papouli, E., Cejka, P. & Jiricny, J. Dependence of the cytotoxicity of DNA-damaging agents on the mismatch repair status of human cells. Cancer Res. 64, 3391–3394 (2004).
Yatabe, Y., Tavaré, S. & Shibata, D. Investigating stem cells in human colon by using methylation patterns. Proc. Natl Acad. Sci. 98, 10839–10844 (2001).
Vogelstein, B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013).
Easwaran, H., Tsai, H.-C. & Baylin, S. B. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell 54, 716–727 (2014).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome — biological and translational implications. Nature. Rev. Cancer 11, 726–734 (2011).
Glasspool, R. M., Teodoridis, J. M. & Brown, R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br. J. Cancer 94, 1087–1092 (2006).
Burrell, R. A. & Swanton, C. The evolution of the unstable cancer genome. Curr. Opin. Genet. Dev. 24, 61–67 (2014).
Cherblanc, F., Chapman-Rothe, N., Brown, R. & Fuchter, M. J. Current limitations and future opportunities for epigenetic therapies. Future Med. Chem. 4, 425–446 (2012).
Kaina, B. & Christmann, M. DNA repair in resistance to alkylating anticancer drugs. Int. J. Clin. Pharmacol. Ther. 40, 354–367 (2002).
Hegi, M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. New Engl. J. Med. 352, 997–1003 (2005).
Wick, W. et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 13, 707–715 (2012).
Malmström, A. et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: the Nordic randomised, phase 3 trial. Lancet Oncol. 13, 916–926 (2012).
Brandes, A. A. et al. O6-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro Oncol. 12, 283–288 (2010).
Felsberg, J. et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int. J. Cancer 129, 659–670 (2011).
Parkinson, J. F. et al. Variation of O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation in serial samples in glioblastoma. J. Neurooncol. 87, 71–78 (2008).
Jung, T.-Y. et al. Changes of the O6-methylguanine-DNA methyltransferase promoter methylation and MGMT protein expression after adjuvant treatment in glioblastoma. Oncol. Rep. 23, 1269–1276 (2010).
Christmann, M. et al. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: a comparative study on astrocytoma and glioblastoma. Int. J. Cancer 127, 2106–2118 (2010).
Park, C.-K. et al. The changes in MGMT promoter methylation status in initial and recurrent glioblastomas. Transl. Oncol. 5, 393–397 (2012).
Zhang, Y.-W. et al. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics http://dx.doi.org/10.4161/epi.28601 (2014).
Plumb, J. A., Strathdee, G., Sludden, J., Kaye, S. B. & Brown, R. Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 60, 6039–6044 (2000).
Chang, X. et al. Identification of hypermethylated genes associated with cisplatin resistance in human cancers. Cancer Res. 70, 2870–2879 (2010).
Strathdee, G., MacKean, M., Illand, M. & Brown, R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene 18, 2335–2341 (1999).
Gifford, G., Paul, J., Vasey, P. A., Kaye, S. B. & Brown, R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin. Cancer Res. 10, 4420–4426 (2004).
McShane, L. M. et al. Reporting recommendations for tumor marker prognostic studies. J. Clin. Oncol. 23, 9067–9072 (2005).
Balko, J. M. et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nature Med. 18, 1052–1059 (2012).
Watanabe, Y. et al. A change in promoter methylation of hMLH1 is a cause of acquired resistance to platinum-based chemotherapy in epithelial ovarian cancer. Anticancer Res. 27, 1449–1452 (2007).
Holohan, C., Van Schaeybroeck, S., Longley, D. B. & Johnston, P. G. Cancer drug resistance: an evolving paradigm. Nature Rev. Cancer 13, 714–726 (2013).
Monsma, D. J. et al. Using a rhabdomyosarcoma patient-derived xenograft to examine precision medicine approaches and model acquired resistance. Pediatr. Blood Cancer 61, 1570–1577 (2014).
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014).
Mendenhall, E. M. et al. Locus-specific editing of histone modifications at endogenous enhancers. Nature Biotech. 31, 1133–1136 (2013).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Beck, B. & Blanpain, C. Unravelling cancer stem cell potential. Nature Rev. Cancer 13, 727–738 (2013).
Shlush, L. I. et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 506, 328–333 (2014).
Rizzo, S. et al. Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2. Mol. Cancer Ther. 10, 325–335 (2011).
Meacham, C. E. & Morrison, S. J. Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337 (2013).
Anderson, K. et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469, 356–361 (2011).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Mullighan, C. G. et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 322, 1377–1380 (2008).
Sharma, S. V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010).
Knoechel, B. et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nature Genet. 46, 364–370 (2014).
Gupta, P. B. et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644 (2011).
Chaffer, C. L. et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl Acad. Sci. 108, 7950–7955 (2011).
Schlesinger, Y. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature Genet. 39, 232–236 (2006).
Ohm, J. E. et al. A stem cell–like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature Genet. 39, 237–242 (2007).
Widschwendter, M. et al. Epigenetic stem cell signature in cancer. Nature Genet. 39, 157–158 (2007).
Easwaran, H. et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome Res. 22, 837–849 (2012).
Zhuang, J. et al. The dynamics and prognostic potential of DNA methylation changes at stem cell gene loci in women's cancer. PLoS Genet. 8, e1002517 (2012).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Azuara, V. et al. Chromatin signatures of pluripotent cell lines. Nature Cell Biol. 8, 532–538 (2006).
Chapman-Rothe, N. et al. Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high grade serous ovarian cancer that distinguish malignant, tumour sustaining and chemo-resistant ovarian tumour cells. Oncogene 19, 4586–4592 (2012).
Chaffer, C. L. et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 154, 61–74 (2013).
Jansen, M. P. et al. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer Res. 73, 6632–6641 (2013).
Magnani, L. et al. Genome-wide reprogramming of the chromatin landscape underlies endocrine therapy resistance in breast cancer. Proc. Natl Acad. Sci. 110, 1490–1499 (2013).
Verma, S. K. et al. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med. Chem. Lett. 3, 1091–1096 (2012).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Knutson, S. K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature Chem. Biol. 8, 890–896 (2012).
Qi, W. et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl Acad. Sci. 109, 21360–21365 (2012).
Konze, K. D. et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem. Biol. 8, 1324–1334 (2013).
Garapaty-Rao, S. et al. Identification of EZH2 and EZH1 small molecule inhibitors with selective impact on diffuse large B cell lymphoma cell growth. Chem. Biol. 20, 1329–1339 (2013).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Bracken, A. P. et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22, 5323–5335 (2003).
Li, H., Cai, Q., Godwin, A. K. & Zhang, R. Enhancer of zeste homolog 2 promotes the proliferation and invasion of epithelial ovarian cancer cells. Mol. Cancer Res. 8, 1610–1618 (2010).
Knutson, S. K. et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl Acad. Sci. 110, 7922–7927 (2013).
Thomas, A. et al. Drug-induced apoptosis in B cell chronic lymphocytic leukemia: relationship between p53 gene mutation and bcl-2/bax proteins in drug resistance. Oncogene 12, 1055–1062 (1996).
Perego, P. et al. Association between cisplatin resistance and mutation of p53 gene and reduced BAX expression in ovarian carcinoma cell systems. Cancer Res. 56, 556–562 (1996).
Senisterra, G. et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem. J. 449, 151–159 (2013).
Chen, Q., Van der Sluis, P. C., Boulware, D., Hazlehurst, L. A. & Dalton, W. S. The FANC/BRCA pathway is involved in melphalan induced DNA interstrand crosslink repair and accounts for melphalan resistance in multiple myeloma cells. Blood 106, 698–705 (2005).
Edwards, S. L. et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111–1115 (2008).
Kitange, G. J. et al. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol. 11, 281–291 (2009).
Zeller, C. & Brown, R. Therapeutic modulation of epigenetic drivers of drug resistance in ovarian cancer. Ther. Adv. Med. Oncol. 2, 319–329 (2010).
Matei, D. et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 72, 2197–2205 (2012).
Glasspool, R. et al. A randomised, Phase II trial of the DNA-hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in combination with carboplatin versus carboplatin alone in patients with recurrent, partially platinum-sensitive ovarian cancer. Br. J. Cancer 110, 1923–1929 (2014).
Fan, H. et al. Low-dose decitabine-based chemoimmunotherapy for patients with refractory advanced solid tumors: a Phase I/II report. J. Immunol. Res. 2014, 371087 (2014).
Appleton, K. et al. Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J. Clin. Oncol. 25, 4603–4609 (2007).
Chapman-Rothe, N. & Brown, R. Approaches to target the genome and its epigenome in cancer. Future Med. Chem. 1, 1481–1495 (2009).
Kitange, G. J. et al. Inhibition of histone deacetylation potentiates the evolution of acquired temozolomide resistance linked to MGMT upregulation in glioblastoma xenografts. Clin. Cancer Res. 18, 4070–4079 (2012).
Li, S. et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 4, 1116–1130 (2013).
Topp, M. D. et al. Molecular correlates of platinum response in human high-grade serous ovarian cancer patient-derived xenografts. Mol. Oncol. 8, 656–668 (2014).
Jaspers, J. E. et al. Loss of 53BP1 causes PARP inhibitor resistance in BRCA1-mutated mouse mammary tumors. Cancer Discov. 3, 68–81 (2013).
O'Hagan, H. M. et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 20, 606–619 (2011).
Bauer, S. et al. Phase I study of panobinostat and imatinib in patients with treatment-refractory metastatic gastrointestinal stromal tumors. Br. J. Cancer 110, 1155–1162 (2014).
Garrido-Laguna, I. et al. A Phase I/II study of decitabine in combination with panitumumab in patients with wild-type (wt) KRAS metastatic colorectal cancer. Invest. New Drugs 31, 1257–1264 (2013).
Falchook, G. S. et al. Methylation and histone deacetylase inhibition in combination with platinum treatment in patients with advanced malignancies. Invest. New Drugs 31, 1192–1200 (2013).
Fu, S. et al. Phase 1b-2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer 117, 1661–1669 (2011).
Sonpavde, G. et al. Azacitidine favorably modulates PSA kinetics correlating with plasma DNA LINE-1 hypomethylation in men with chemonaïve castration-resistant prostate cancer. Urol. Oncol. 29, 682–689 (2011).
Munster, P. et al. A Phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br. J. Cancer 104, 1828–1835 (2011).
Dizon, D. S. et al. A Phase II evaluation of belinostat and carboplatin in the treatment of recurrent or persistent platinum-resistant ovarian, fallopian tube, or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol. Oncol. 125, 367–371 (2012).
Dizon, D. S. et al. Phase II activity of belinostat (PXD-101), carboplatin, and paclitaxel in women with previously treated ovarian cancer. Int. J. Gynecol. Cancer 22, 979–986 (2012).
Acknowledgements
The authors thank S. Kaye and E. Loomis for comments on the manuscript. The authors' research on epigenetics of drug resistance is supported by grants from Cancer Research UK (A13086) and Ovarian Cancer Action, London, UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Grants from Cancer Research UK and Ovarian Cancer Action, which is acknowledged (R.B.). Patent on histone methyltransferase inhibitors (R.B.). E.C., L.M., C.S.W.-B and J.B. declare no competing interests.
Related links
FURTHER INFORMATION
PowerPoint slides
Supplementary information
41568_2014_BFnrc3819_MOESM11_ESM.pdf
Supplementary information S1 | Systematic review of the published literature on epigenetic mechanisms involved in drug resistance in clinical studies (PDF 369 kb)
Rights and permissions
About this article
Cite this article
Brown, R., Curry, E., Magnani, L. et al. Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer 14, 747–753 (2014). https://doi.org/10.1038/nrc3819
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrc3819
This article is cited by
-
Epigenetic-based combination therapy and liposomal codelivery overcomes osimertinib-resistant NSCLC via repolarizing tumor-associated macrophages
Acta Pharmacologica Sinica (2024)
-
Protein degradation: expanding the toolbox to restrain cancer drug resistance
Journal of Hematology & Oncology (2023)
-
Multiplexed, single-molecule, epigenetic analysis of plasma-isolated nucleosomes for cancer diagnostics
Nature Biotechnology (2023)
-
A comparison of mutation and amplification-driven resistance mechanisms and their impacts on tumor recurrence
Journal of Mathematical Biology (2023)
-
Detection of metabolic adaptation in a triple-negative breast cancer animal model with [18F]choline-PET imaging as a surrogate for drug resistance
European Journal of Nuclear Medicine and Molecular Imaging (2023)