Abstract
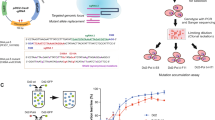
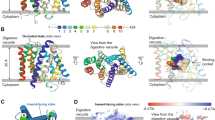
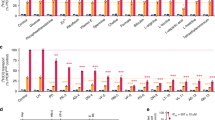
A molecular understanding of drug resistance mechanisms enables surveillance of the effectiveness of new antimicrobial therapies during development and deployment in the field. We used conventional drug resistance selection as well as a regime of limiting dilution at early stages of drug treatment to probe two antimalarial imidazolopiperazines, KAF156 and GNF179. The latter approach permits the isolation of low-fitness mutants that might otherwise be out-competed during selection. Whole-genome sequencing of 24 independently derived resistant Plasmodium falciparum clones revealed four parasites with mutations in the known cyclic amine resistance locus (pfcarl) and a further 20 with mutations in two previously unreported P. falciparum drug resistance genes, an acetyl-CoA transporter (pfact) and a UDP-galactose transporter (pfugt). Mutations were validated both in vitro by CRISPR editing in P. falciparum and in vivo by evolution of resistant Plasmodium berghei mutants. Both PfACT and PfUGT were localized to the endoplasmic reticulum by fluorescence microscopy. As mutations in pfact and pfugt conveyed resistance against additional unrelated chemical scaffolds, these genes are probably involved in broad mechanisms of antimalarial drug resistance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
WHO. World Malaria Report 2015 1–280 (WHO Press, 2015).
Dondorp, A. M. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361, 455–467 (2009).
Ashley, E. A. et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 371, 411–423 (2014).
Nagle, A. et al. Imidazolopiperazines: lead optimization of the second-generation antimalarial agents. J. Med. Chem. 55, 4244–4273 (2012).
Wu, T. et al. Imidazolopiperazines: hit to lead optimization of new antimalarial agents. J. Med. Chem. 54, 5116–5130 (2011).
Leong, F. J. et al. A first-in-human randomized, double-blind, placebo-controlled, single- and multiple-ascending oral dose study of novel imidazolopiperazine KAF156 to assess its safety, tolerability, and pharmacokinetics in healthy adult volunteers. Antimicrob. Agents Chemother. 58, 6437–6443 (2014).
Kuhen, K. L. et al. KAF156 is an antimalarial clinical candidate with potential for use in prophylaxis, treatment, and prevention of disease transmission. Antimicrob. Agents Chemother. 58, 5060–5067 (2014).
Ding, X. C., Ubben, D. & Wells, T. N. A framework for assessing the risk of resistance for anti-malarials in development. Malaria J. 11, 292 (2012).
Jimenez-Diaz, M. B. et al. (+)-SJ733, a clinical candidate for malaria that acts through ATP4 to induce rapid host-mediated clearance of Plasmodium. Proc. Natl Acad. Sci. USA 111, E5455–E5462 (2014).
Baragana, B. et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 522, 315–320 (2015).
Flannery, E. L., Fidock, D. A. & Winzeler, E. A. Using genetic methods to define the targets of compounds with antimalarial activity. J. Med. Chem. 56, 7761–7771 (2013).
Meister, S. et al. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science 334, 1372–1377 (2011).
LaMonte, G. et al. Mutations in the Plasmodium falciparum Cyclic Amine Resistance Locus (PfCARL) confer multidrug resistance. mBio 7, e00696-16 (2016).
Martin, R. E., Henry, R. I., Abbey, J. L., Clements, J. D. & Kirk, K. The ‘permeome’ of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 6, R26 (2005).
Valderramos, S. G. & Fiddock, D. A. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 27, 594–601 (2006).
Ecker, A., Lehane, A. M., Clain, J. & Fidock, D. A. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 28, 504–514 (2012).
Johnson, J. D. et al. Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob. Agents Chemother. 51, 1926–1933 (2007).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Yan, N. Structural biology of the major facilitator superfamily transporters. Annu. Rev. Biophys. 44, 257–283 (2015).
Pedersen, B. P. et al. Crystal structure of a eukaryotic phosphate transporter. Nature 496, 533–536 (2013).
Quistgaard, E. M., Low, C., Guettou, F. & Nordlund, P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat. Rev. Mol. Cell Biol. 17, 123–132 (2016).
Lee, M. C. & Fidock, D. A. CRISPR-mediated genome editing of Plasmodium falciparum malaria parasites. Genome Med. 6, 63 (2014).
Ginsburg, H. Malaria Parasite Metabolic Pathways (2015), http://mpmp.huji.ac.il/maps/ERGolgiVacuole.html
Lee, M. C., Moura, P. A., Miller, E. A. & Fidock, D. A. Plasmodium falciparum Sec24 marks transitional ER that exports a model cargo via a diacidic motif. Mol. Microbiol. 68, 1535–1546 (2008).
Hayakawa, Y. et al. Structure of tyroscherin, an antitumor antibiotic against IGF-1-dependent cells from Pseudallescheria sp. J. Antibiot. (Tokyo) 57, 634–638 (2004).
Hediger, M. A. et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins. Pflugers Arch. 447, 465–468 (2004).
Prasad, R. & Rawal, M. K. Efflux pump proteins in antifungal resistance. Front. Pharmacol. 5, 202 (2014).
Kumar, S. et al. Bacterial multidrug efflux pumps of the major facilitator superfamily as targets for modulation. Infect. Disord. Drug Targets 16, 28–43 (2016).
Dos Santos, S. C., Teixeira, M. C., Dias, P. J. & Sa-Correia, I. MFS transporters required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast understanding their physiological function through post-genomic approaches. Front. Physiol. 5, 180 (2014).
Aurrecoechea, C. et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543 (2009).
Perlin, M. H., Andrews, J. & Toh, S. S. Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. Adv. Genet. 85, 201–253 (2014).
Jack, D. L., Yang, N. M. & Saier, M. H. Jr. The drug/metabolite transporter superfamily. Eur. J. Biochem. 268, 3620–3639 (2001).
Tran, C. V. & Saier, M. H. Jr. The principal chloroquine resistance protein of Plasmodium falciparum is a member of the drug/metabolite transporter superfamily. Microbiology 150, 1–3 (2004).
Martin, R. E. & Kirk, K. The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol. Biol. Evol. 21, 1938–1949 (2004).
Ng, B. G. et al. Mosaicism of the UDP-galactose transporter SLC35A2 causes a congenital disorder of glycosylation. Am. J. Hum. Gent. 92, 632–636 (2013).
Sprong, H. et al. Association of the Golgi UDP-galactose transporter with UDP-galactose:ceramide galactosyltransferase allows UDP-galactose import in the endoplasmic reticulum. Mol. Biol. Cell 14, 3482–3493 (2003).
Dorre, K. et al. A new case of UDP-galactose transporter deficiency (SLC35A2-CDG): molecular basis, clinical phenotype, and therapeutic approach. J. Inherit. Metab. Dis. 38, 931–940 (2015).
Kanamori, A. et al. Expression cloning and characterization of a cDNA encoding a novel membrane protein required for the formation of O-acetylated ganglioside: a putative acetyl-CoA transporter. Proc. Natl Acad. Sci. USA 94, 2897–2902 (1997).
Varki, A. & Diaz, S. The transport and utilization of acetyl coenzyme A by rat liver Golgi vesicles. O-acetylated sialic acids are a major product. J. Biol. Chem. 260, 6600–6608 (1985).
Jonas, M. C., Pehar, M. & Puglielli, L. AT-1 is the ER membrane acetyl-CoA transporter and is essential for cell viability. J. Cell Sci. 123, 3378–3388 (2010).
Hirabayashi, Y., Nomura, K. H. & Nomura, K. The acetyl-CoA transporter family SLC33. Mol. Aspects Med. 34, 586–589 (2013).
McNamara, C. W. et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature 504, 248–253 (2013).
Walch-Solimena, C. & Novick, P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1, 523–525 (1999).
Kruger, T., Sanchez, C. P. & Lanzer, M. Complementation of Saccharomyces cerevisiae Pik1ts by a phosphatidylinositol 4-kinase from Plasmodium falciparum. Mol. Biochem. Parasitol. 172, 149–151 (2010).
Roemer, T. et al. Confronting the challenges of natural product-based antifungal discovery. Chem. Biol. 18, 148–164 (2011).
Goodman, C. D. et al. Parasites resistant to the antimalarial atovaquone fail to transmit by mosquitoes. Science 352, 349–353 (2016).
Trager, W. & Jensen, J. B. Human malaria parasites in continuous culture. Science 193, 673–675 (1979).
Fidock, D. A., Rosenthal, P. J., Croft, S. L., Brun, R. & Nwaka, S. Antimalarial drug discovery: efficacy models for compound screening. Nat. Rev. Drug Discov. 3, 509–520 (2004).
Kariuki, M. M. et al. Plasmodium falciparum: purification of the various gametocyte developmental stages from in vitro-cultivated parasites. Am. J. Trop. Med. Hyg. 59, 505–508 (1998).
Manary, M. J. et al. Identification of pathogen genomic variants through an integrated pipeline. BMC Bioinformatics 15, 63 (2014).
Ng, C. L. et al. CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Mol. Microbiol. 101, 381–393 (2016).
Malleret, B. et al. A rapid and robust tri-color flow cytometry assay for monitoring malaria parasite development. Sci. Rep. 1, 118 (2011).
Nkrumah, L. J. et al. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nat. Methods 3, 615–621 (2006).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Fiser, A. & Sali, A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 (2003).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E. A. & Bacon, D. J. Raster3D Photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Acknowledgements
M.Y.L. is supported by the Economic Development Board—Industrial Postgraduate Programme (EDB-IPP) scholarship. G.L. is supported by an A.P. Giannini Post-Doctoral Fellowship. Work at UCSD was supported by grants from the National Institutes of Health (NIH; R01 AI090141 and R01 AI103058) to E.A.W. D.A.F. acknowledges support from the Medicines for Malaria Venture. B.M. and L.R. are supported by the Singapore Immunology Network under the Agency for Science, Technology and Research (A*STAR, Singapore). R.W. is a Research Associate at the National Fund for Scientific Research FNRS-FRS (Belgium). The authors thank C. Jensen (Leiden University Medical Center, Netherlands) for the donation of the P. berghei ANKA strain.
Author information
Authors and Affiliations
Contributions
M.Y.-X.L., G.L., M.C.S.L., E.A.W. and P.B. designed the experiments. M.Y.-X.L., G.L., M.C.S.L., C.R., B.H.T., V.C., B.F.T., A.C., M.N., B.M., E.D.C. and L.L. performed the experiments. Modelling work was performed by R.W. M.Y.-X.L., G.L., M.C.S.L., C.R., V.C., M.N., E.D.C. and P.G. analysed the data. G.M.C.B., P.C.-L.H., L.R., D.A.F. and T.T.D. contributed support. M.Y.-X.L., G.L., M.C.S.L., B.K.S.Y., D.A.F., E.A.W. and P.B. wrote and proofread the manuscript. E.A.W. and P.B. gave technical support and conceptual advice. The manuscript was edited by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Tables 1–3, Supplementary Figures 1–4 (PDF 1035 kb)
Rights and permissions
About this article
Cite this article
Lim, MX., LaMonte, G., Lee, M. et al. UDP-galactose and acetyl-CoA transporters as Plasmodium multidrug resistance genes. Nat Microbiol 1, 16166 (2016). https://doi.org/10.1038/nmicrobiol.2016.166
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.166
This article is cited by
-
Antimalarial drug discovery: progress and approaches
Nature Reviews Drug Discovery (2023)
-
Creation and preclinical evaluation of genetically attenuated malaria parasites arresting growth late in the liver
npj Vaccines (2022)
-
A heat-shock response regulated by the PfAP2-HS transcription factor protects human malaria parasites from febrile temperatures
Nature Microbiology (2021)
-
Defining multiplicity of vector uptake in transfected Plasmodium parasites
Scientific Reports (2020)
-
Pan-active imidazolopiperazine antimalarials target the Plasmodium falciparum intracellular secretory pathway
Nature Communications (2020)