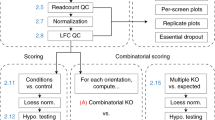
Abstract
RNA interference (RNAi) has become a powerful technique for reverse genetics and drug discovery, and in both of these areas large-scale high-throughput RNAi screens are commonly performed. The statistical techniques used to analyze these screens are frequently borrowed directly from small-molecule screening; however, small-molecule and RNAi data characteristics differ in meaningful ways. We examine the similarities and differences between RNAi and small-molecule screens, highlighting particular characteristics of RNAi screen data that must be addressed during analysis. Additionally, we provide guidance on selection of analysis techniques in the context of a sample workflow.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Boutros, M. & Ahringer, J. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 9, 554–566 (2008).
Perrimon, N. & Mathey-Prevot, B. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics 175, 7–16 (2007).
Jackson, A.L. et al. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21, 635–637 (2003).
Birmingham, A. et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204 (2006).
Jackson, A.L. et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187 (2006).
Echeverri, C.J. et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods 3, 777–779 (2006).
Chatterjee-Kishore, M. From genome to phenome–RNAi library screening and hit characterization using signaling pathway analysis. Curr. Opin. Drug Discov. Devel. 9, 231–239 (2006).
Ramadan, N., Flockhart, I., Booker, M., Perrimon, N. & Mathey-Prevot, B. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat. Protoc. 2, 2245–2264 (2007).
Malo, N., Hanley, J.A., Cerquozzi, S., Pelletier, J. & Nadon, R. Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 24, 167–175 (2006). A comprehensive but accessible introduction to the small-molecule screening analysis techniques, which form the starting point for most RNAi screen analyses.
Zhang, X.H.D. Novel analytic criteria and effective plate designs for quality control in genome-scale RNAi screens. J. Biomol. Screen. 13, 363–377 (2008). This article describes the use of SSMD for quality assessment and suggests appropriate thresholds, as well as provides an in-depth discussion of plate layout issues.
Zhang, X.H.D. et al. Integrating experimental and analytic approaches to improve data quality in genome-wide RNAi screens. J. Biomol. Screen. 13, 378–389 (2008).
Zhang, X.H.D. & Heyse, J.F. Determination of sample size in genome-scale RNAi screens. Bioinformatics 25, 841–844 (2009).
Gunter, B., Brideau, C., Pikounis, B. & Liaw, A. Statistical and graphical methods for quality control determination of high-throughput screening data. J. Biomol. Screen. 8, 624–633 (2003).
Wiles, A.M., Ravi, D., Bhavani, S. & Bishop, A.J. An analysis of normalization methods for Drosophila RNAi genomic screens and development of a robust validation scheme. J. Biomol. Screen. 13, 777–784 (2008).
Brideau, C., Gunter, B., Pikounis, B. & Liaw, A. Improved statistical methods for hit selection in high-throughput screening. J. Biomol. Screen. 8, 634–647 (2003).
Boutros, M., Brás, L.P. & Huber, W. Analysis of cell-based RNAi screens. Genome Biol. 7, R66 (2006).
Zhang J.H., Chung T.D. & Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. B, 4 67–73 (1999).
Zhang, X.H.D. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics 89, 552–561 (2007).
Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 27, 861–874 (2006).
Wagner, E.K. et al. Practical approaches to long oligonucleotide-based DNA microarray: lessons from herpesviruses. Prog. Nucleic Acid Res. Mol. Biol. 71, 445–491 (2002).
Forster, T., Roy, D. & Ghazal, P. Experiments using microarray technology: limitations and standard operating procedures. J. Endocrinol. 178, 195–204 (2003).
Stone, D.J. et al. High-throughput screening by RNA interference: control of two distinct types of variance. Cell Cycle 6, 898–901 (2007).
Ainscow, E. Statistical techniques for handling high content screening data. European Pharmaceutical Review 5, 30–38 (2007).
Bard, F. et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439, 604–607 (2006).
DasGupta, R., Kaykas, A., Moon, R.T. & Perrimon, N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308, 826–833 (2005).
Zhang, X.H.D. et al. Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics 7, 299–309 (2006).
Chung, N. et al. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J. Biomol. Screen. 13, 149–158 (2008). This article describes experimental and computational studies validating the effectiveness of median ± k median absolute deviations as a hit identification approach for RNAi screens.
Müller, P., Kuttenkeuler, D., Gesellchen, V., Zeidler, M.P. & Boutros, M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature 436, 871–875 (2005).
Whitehurst, A.W. et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446, 815–819 (2007).
Manly, K.F., Nettleton, D. & Hwang, J.T. Genomics, prior probability, and statistical tests of multiple hypotheses. Genome Res. 14, 997–1001 (2004).
Zhang, X.H.D. Genome-wide screens for effective siRNAs through assessing the size of siRNA effects. BMC Res. Notes 1, 33 (2008).
Zhang, X.H.D., Marine, S.D. & Ferrer, M. Error rates and powers in genome-scale RNAi screens. J. Biomol. Screen. 14, 230–238 (2009).
Zhang, X.H.D. A new method with flexible and balanced control of false negatives and false positives for hit selection in RNA interference high-throughput screening assays. J. Biomol. Screen. 12, 645–655 (2007).
Zhang, X.H.D. et al. The use of strictly standardized mean difference for hit selection in primary RNA interference high-throughput screening experiments. J. Biomol. Screen. 12, 497–509 (2007).
König, R. et al. A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods 4, 847–849 (2007).
Breitling, R., Armengaud, P., Amtmann, A. & Herzyk, P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92 (2004).
Rieber, N., Knapp, B., Eils, R. & Kaderali, L. RNAither, an automated pipeline for the statistical analysis of high-throughput RNAi screens. Bioinformatics 25, 678–679 (2009).
Zhang, X.H.D. et al. Hit selection with false discovery rate control in genome-scale RNAi screens. Nucleic Acids Res. 36, 4667–4679 (2008).
Acknowledgements
Work at ICCB-Longwood was funded by US National Institutes of Health grants CA078048, AI067751 and AI057159. Funding to P.G. was provided by the Wellcome Trust and the Biotechnology and Biological Sciences Research Council (BBSRC). The Centre for Systems Biology at Edinburgh is a Centre for Integrative Systems Biology (CISB) funded by BBSRC and the Engineering and Physical Sciences Research Council (EPSRC), reference BB/D019621/1. Work at the University of Edinburgh was supported by edikt2 (Scottish Funding Council grant HR04019; http://www.edikt.org/). R.L.B. is supported by Shering-Plough and TIPharma. D.J.D., A.L. and D.K. are supported by Marie Curie MTKD-CT-2005-029798 (European Union FP6).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
A.B. and Q.S. are employed by Thermo Fisher Scientific. T.F. and P.G. are directors of Fios Genomics Ltd. C.J.K. is now employed by Life Technologies.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–2 (PDF 206 kb)
Rights and permissions
About this article
Cite this article
Birmingham, A., Selfors, L., Forster, T. et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods 6, 569–575 (2009). https://doi.org/10.1038/nmeth.1351
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1351
This article is cited by
-
Phenotypic screening in Organ-on-a-Chip systems: a 1537 kinase inhibitor library screen on a 3D angiogenesis assay
Angiogenesis (2024)
-
PHDs-seq: a large-scale phenotypic screening method for drug discovery through parallel multi-readout quantification
Cell Regeneration (2023)
-
Transfer learning for versatile and training free high content screening analyses
Scientific Reports (2023)
-
The function and evolution of a genetic switch controlling sexually dimorphic eye differentiation in honeybees
Nature Communications (2023)
-
A genome-wide RNAi screen for genes important for proliferation of cultured Drosophila cells at low temperature identifies the Ball/VRK protein kinase
Chromosoma (2023)