Abstract
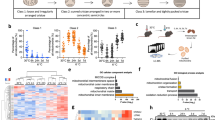
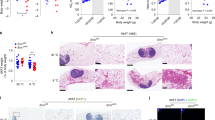
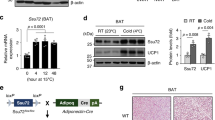
Adipocytes possess remarkable adaptive capacity to respond to nutrient excess, fasting or cold exposure, and they are thus an important cell type for the maintenance of proper metabolic health. Although the endoplasmic reticulum (ER) is a critical organelle for cellular homeostasis, the mechanisms that mediate adaptation of the ER to metabolic challenges in adipocytes are unclear. Here we show that brown adipose tissue (BAT) thermogenic function requires an adaptive increase in proteasomal activity to secure cellular protein quality control, and we identify the ER-localized transcription factor nuclear factor erythroid 2–like 1 (Nfe2l1, also known as Nrf1) as a critical driver of this process. We show that cold adaptation induces Nrf1 in BAT to increase proteasomal activity and that this is crucial for maintaining ER homeostasis and cellular integrity, specifically when the cells are in a state of high thermogenic activity. In mice, under thermogenic conditions, brown-adipocyte-specific deletion of Nfe2l1 (Nrf1) resulted in ER stress, tissue inflammation, markedly diminished mitochondrial function and whitening of the BAT. In mouse models of both genetic and dietary obesity, stimulation of proteasomal activity by exogenously expressing Nrf1 or by treatment with the proteasome activator PA28α in BAT resulted in improved insulin sensitivity. In conclusion, Nrf1 emerges as a novel guardian of brown adipocyte function, providing increased proteometabolic quality control for adapting to cold or to obesity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004).
Bartelt, A. et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205 (2011).
Stanford, K.I. et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J. Clin. Invest. 123, 215–223 (2013).
Berbée, J.F. et al. Brown fat activation reduces hypercholesterolemia and protects from atherosclerosis development. Nat. Commun. 6, 6356 (2015).
Bartelt, A. et al. Thermogenic adipocytes promote HDL turnover and reverse cholesterol transport. Nat. Commun. 8, 15010 (2017).
Harms, M. & Seale, P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 (2013).
Kajimura, S., Spiegelman, B.M. & Seale, P. Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 22, 546–559 (2015).
Gnad, T. et al. Adenosine activates brown adipose tissue and recruits beige adipocytes via A2A receptors. Nature 516, 395–399 (2014).
Rothwell, N.J. & Stock, M.J. A role for brown adipose tissue in diet-induced thermogenesis. Nature 281, 31–35 (1979).
Feldmann, H.M., Golozoubova, V., Cannon, B. & Nedergaard, J. Ucp1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 9, 203–209 (2009).
Shimizu, I. et al. Vascular rarefaction mediates whitening of brown fat in obesity. J. Clin. Invest. 124, 2099–2112 (2014).
Cypess, A.M. et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517 (2009).
Saito, M. et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58, 1526–1531 (2009).
Travers, K.J. et al. Functional and genomic analyses reveal an essential coordination between the unfolded-protein response and ER-associated degradation. Cell 101, 249–258 (2000).
Friedlander, R., Jarosch, E., Urban, J., Volkwein, C. & Sommer, T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2, 379–384 (2000).
Goldberg, A.L. Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 (2003).
Ron, D. & Walter, P. Signal integration in the endoplasmic reticulum unfolded-protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 (2007).
Hotamisligil, G.S. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140, 900–917 (2010).
Yang, L. et al. METABOLISM. S-nitrosylation links obesity-associated inflammation to endoplasmic reticulum dysfunction. Science 349, 500–506 (2015).
Otoda, T. et al. Proteasome dysfunction mediates obesity-induced endoplasmic reticulum stress and insulin resistance in the liver. Diabetes 62, 811–824 (2013).
Wikstrom, J.D. et al. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. EMBO J. 33, 418–436 (2014).
Arruda, A.P. et al. Chronic enrichment of hepatic endoplasmic reticulum–mitochondria contact leads to mitochondrial dysfunction in obesity. Nat. Med. 20, 1427–1435 (2014).
Gregor, M.F. et al. The role of adipocyte XBP1 in metabolic regulation during lactation. Cell Rep. 3, 1430–1439 (2013).
Wang, W. & Chan, J.Y. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J. Biol. Chem. 281, 19676–19687 (2006).
Cannon, B. & Nedergaard, J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. J. Exp. Biol. 214, 242–253 (2011).
Adams, J. et al. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 59, 2615–2622 (1999).
Sha, Z. & Goldberg, A.L. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Curr. Biol. 24, 1573–1583 (2014).
Arastu-Kapur, S. et al. Nonproteasomal targets of the proteasome inhibitors bortezomib and carfilzomib: a link to clinical adverse events. Clin. Cancer Res. 17, 2734–2743 (2011).
Fu, S. et al. Phenotypic assays identify azoramide as a small-molecule modulator of the unfolded-protein response with antidiabetic activity. Sci. Transl. Med. 7, 292ra98 (2015).
Ozcan, U. et al. Endoplasmic reticulum stress links obesity, insulin action and type 2 diabetes. Science 306, 457–461 (2004).
Steffen, J., Seeger, M., Koch, A. & Krüger, E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 40, 147–158 (2010).
Radhakrishnan, S.K. et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28 (2010).
Kahn, N.W., Rea, S.L., Moyle, S., Kell, A. & Johnson, T.E. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. Biochem. J. 409, 205–213 (2008).
Xue, R. et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat. Med. 21, 760–768 (2015).
Bartelt, A. & Heeren, J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 10, 24–36 (2014).
Peirce, V., —, S. & Vidal-Puig, A. The different shades of fat. Nature 510, 76–83 (2014).
Rosen, E.D. & Spiegelman, B.M. What we talk about when we talk about fat. Cell 156, 20–44 (2014).
Bruns, O.T. et al. Next-generation in vivo optical imaging with shortwave infrared quantum dots. Nat. Biomed. Eng. 1 (2017).
Hershko, A. & Ciechanover, A. The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807 (1992).
Kulathu, Y. & Komander, D. Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13, 508–523 (2012).
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011).
Wagner, S.A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics 10, M111.013284 (2011).
Mootha, V.K. et al. Integrated analysis of protein composition, tissue diversity and gene regulation in mouse mitochondria. Cell 115, 629–640 (2003).
Clarke, K.J. et al. A role for ubiquitinylation and the cytosolic proteasome in turnover of mitochondrial uncoupling protein 1 (UCP1). Biochim. Biophys. Acta 1817, 1759–1767 (2012).
Mootha, V.K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Kamburov, A. et al. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 39, D712–D717 (2011).
Xu, H. et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 112, 1821–1830 (2003).
Weisberg, S.P. et al. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112, 1796–1808 (2003).
Lumeng, C.N., Bodzin, J.L. & Saltiel, A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117, 175–184 (2007).
Hotamisligil, G.S., Shargill, N.S. & Spiegelman, B.M. Adipose expression of tumor necrosis factor–α: direct role in obesity-linked insulin resistance. Science 259, 87–91 (1993).
Kim, J.Y. et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest. 117, 2621–2637 (2007).
Li, J. et al. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia–reperfusion injury in mice. J. Clin. Invest. 121, 3689–3700 (2011).
Zhang, Y. et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 513, 440–443 (2014).
Lee, C.S., Ho, D.V. & Chan, J.Y. Nuclear factor–erythroid-2-related factor 1 regulates expression of proteasome genes in hepatocytes and protects against endoplasmic reticulum stress and steatosis in mice. FEBS J. 280, 3609–3620 (2013).
Wahl, S. et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 541, 81–86 (2017).
Zhang, Y. & Xiang, Y. Molecular and cellular basis for the unique functioning of Nrf1, an indispensable transcription factor for maintaining cell homoeostasis and organ integrity. Biochem. J. 473, 961–1000 (2016).
Widenmaier, S.B. et al. NRF1 is an ER membrane sensor that is central to cholesterol homeostasis. Cell 171, 1094–1109 (2017).
Tsujita, T. et al. Discovery of an NRF1-specific inducer from a large-scale chemical library using a direct NRF1–protein monitoring system. Genes Cells 20, 563–577 (2015).
Li, F., Gao, B., Dong, H., Shi, J. & Fang, D. Icariin induces synoviolin expression through NFE2L1 to protect neurons from ER-stress-induced apoptosis. PLoS One 10, e0119955 (2015).
Kong, X. et al. IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell 158, 69–83 (2014).
Tschöp, M.H. et al. A guide to analysis of mouse energy metabolism. Nat. Methods 9, 57–63 (2011).
Cypess, A.M. et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 19, 635–639 (2013).
Rodbell, M. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J. Biol. Chem. 239, 375–380 (1964).
Brand, M.D. & Nicholls, D.G. Assessing mitochondrial dysfunction in cells. Biochem. J. 435, 297–312 (2011).
Cannon, B. & Nedergaard, J. Studies of thermogenesis and mitochondrial function in adipose tissues. Methods Mol. Biol. 456, 109–121 (2008).
Orr, A.L., Quinlan, C.L., Perevoshchikova, I.V. & Brand, M.D. A refined analysis of superoxide production by mitochondrial sn-glycerol 3-phosphate dehydrogenase. J. Biol. Chem. 287, 42921–42935 (2012).
Haemmerle, G. et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737 (2006).
Luo, J. et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2, 1236–1247 (2007).
Acknowledgements
We are grateful to K. Claiborn for her expert editorial input, M. McGrath for his support of the project, M. Yilmaz, Y. Lee and V. Byles for assistance with experiments, K. Seedorf, P. Delerive and Servier for their continuous support of the project, R. Bronson and the Harvard Medical School Rodent Histopathology Core, M. Ericson and the Harvard Medical School Electron Microscopy Facility, N. Lupoli and the Harvard T.H. Chan School of Public Health Trace Metals Laboratory, and R. Kunz and the Thermo Fisher Scientific Center for Multiplexed Proteomics at Harvard Medical School and the Harvard T.H. Chan School of Public Health Animal Facility team for their support of the project. We acknowledge the prosperous scientific environment in the Hotamisligil lab and thank all lab members for their critical input and discussions. We are grateful to X. Wang (University of South Dakota) for generously sharing the PA28α-expressing adenovirus, E. Rosen (Beth Israel Deaconess Medical Center) for providing us with the Ucp1–Cre and the Adipoq–Cre mouse models and T. Iwawaki (Gunma University) for the conditional Ern1-deletion mouse model. A.B. was supported by a Deutsche Forschungsgemeinschaft Research Fellowship (BA 4925/1-1). S.B.W. was supported by a Canadian Institutes of Health Research fellowship. C.S. was supported by a University Medical Center Hamburg–Eppendorf MD/PhD program fellowship. R.L.S.G. was supported by the Barth Syndrome Foundation. This work has received support in part from the US National Institutes of Health (NIH) through the Laser Biomedical Research Center grants P41-EB015871 (M.G.B.) and 5-U54-CA151884 (M.G.B.), by the National Science Foundation grant ECCS-1449291 (D.F. and M.G.B.) and by the Massachusetts Institute of Technology through the Institute for Soldier Nanotechnologies grant W911NF-13-D-0001 (M.G.B.). D.F. was supported by a fellowship of the Boehringer Ingelheim Fonds. O.T.B. was supported by a European Molecular Biology Organization Long-term Fellowship. Y.-H.T. and L.O.L. were supported in part by NIH grants R01DK077097, R01DK102898 and P30DK036836. M.D.L. was supported by NIH grants T32DK007260, F32DK102320 and K01DK111714. A.P.A. was supported by the Pew Foundation. We apologize to colleagues whose work we could not cite due to space limitations. The study was supported by an industry-sponsored research agreement between Harvard University and Servier.
Author information
Authors and Affiliations
Contributions
A.B. and S.B.W. designed the study, were involved in all aspects of the experiments, analyzed all data and wrote the manuscript; C.S. and K.J. performed in vitro experiments; R.L.S.G. performed mitochondrial isolations and Seahorse experiments; K.E. performed flow cytometry experiments; A.W.F. performed immunohistochemistry; G.P., N.A.S. and T.B.N. were involved in animal experiments; O.T.B., D.F. and M.G.B. developed quantum dots and performed SWIR imaging; L.O.L. performed thermal imaging; M.D.L. and Y.-H.T. performed bioinformatics analyses; A.P.A. and K.E.I. helped with the design of the study and animal experiments; and G.S.H. supervised the design and execution of the study, interpreted the results and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
Supplementary Figures 1–13 and Supplementary Methods (PDF 5513 kb)
Supplementary Table 1
The brown fat ubiquitome (XLSX 3031 kb)
Supplementary Table 2
The Differential brown fat ubiquitome (XLSX 1589 kb)
Supplementary Table 3
Pathway analysis of hyperubiquitinated proteins (XLSX 67 kb)
Supplementary Table 4
Plasma parameters in high-fat diet-fed wild-type and Nfe2l1ΔBAT mice (XLSX 40 kb)
Supplementary Table 5
Additional information on adenoviral experiments (XLSX 41 kb)
Supplementary Table 6
Primers and antibodies information (XLSX 43 kb)
41591_2018_BFnm4481_MOESM9_ESM.avi
SWIR-QD-lipoprotein uptake imaging of a cold-exposed wild-type mouse: A representative SWIR movie of a cold-exposed control wild-type mouse. After tail vein injection, lipoproteins labeled with short-wave infrared (SWIR) quantum dots (QD) are taken up into liver, spleen and brown adipose tissue. (AVI 14278 kb)
41591_2018_BFnm4481_MOESM10_ESM.avi
SWIR-QD-lipoprotein uptake imaging of a cold-exposed Nfe2lΔBAT mouse: A representative SWIR movie of a cold-exposed control Nfe2lΔBAT mouse. After tail vein injection, lipoproteins labeled with short-wave infrared (SWIR) quantum dots (QD) are taken up into liver, spleen but not into brown adipose tissue. (AVI 13941 kb)
Rights and permissions
About this article
Cite this article
Bartelt, A., Widenmaier, S., Schlein, C. et al. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nat Med 24, 292–303 (2018). https://doi.org/10.1038/nm.4481
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4481
This article is cited by
-
Cardiac proteostasis in obesity and cardiovascular disease
Herz (2024)
-
Ssu72 phosphatase is essential for thermogenic adaptation by regulating cytosolic translation
Nature Communications (2023)
-
Brown fat has a sweet tooth
Nature Metabolism (2023)
-
Integrated analysis of microRNAs, circular RNAs, long non-coding RNAs, and mRNAs revealed competing endogenous RNA networks involved in brown adipose tissue whitening in rabbits
BMC Genomics (2022)
-
Dual roles of mTORC1-dependent activation of the ubiquitin-proteasome system in muscle proteostasis
Communications Biology (2022)