Abstract
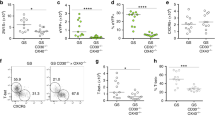
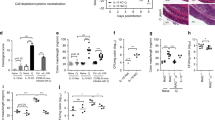
Defense against attaching-and-effacing bacteria requires the sequential generation of interleukin 23 (IL-23) and IL-22 to induce protective mucosal responses. Although CD4+ and NKp46+ innate lymphoid cells (ILCs) are the critical source of IL-22 during infection, the precise source of IL-23 is unclear. We used genetic techniques to deplete mice of specific subsets of classical dendritic cells (cDCs) and analyzed immunity to the attaching-and-effacing pathogen Citrobacter rodentium. We found that the signaling receptor Notch2 controlled the terminal stage of cDC differentiation. Notch2-dependent intestinal CD11b+ cDCs were an obligate source of IL-23 required for survival after infection with C. rodentium, but CD103+ cDCs dependent on the transcription factor Batf3 were not. Our results demonstrate a nonredundant function for CD11b+ cDCs in the response to pathogens in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Mangan, P.R. et al. Transforming growth factor-β induces development of the TH17 lineage. Nature 441, 231–234 (2006).
Zheng, Y. et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 (2008).
Spits, H. & Di Santo, J.P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12, 21–27 (2011).
Colonna, M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity 31, 15–23 (2009).
Sonnenberg, G.F. et al. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134 (2011).
Zheng, Y. et al. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature 445, 648–651 (2007).
Mundy, R. et al. Citrobacter rodentium of mice and man. Cell. Microbiol. 7, 1697–1706 (2005).
Cella, M. et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457, 722–725 (2009).
Eberl, G. et al. An essential function for the nuclear receptor RORγ(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5, 64–73 (2004).
Sanos, S.L. et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10, 83–91 (2009).
Tumanov, A.V. et al. Lymphotoxin controls the IL-22 protection pathway in gut innate lymphoid cells during mucosal pathogen challenge. Cell Host Microbe 10, 44–53 (2011).
Manta, C. et al. CX3CR1+ macrophages support IL-22 production by innate lymphoid cells during infection with Citrobacter rodentium. Mucosal Immunol. 6, 177–188 (2012).
Kinnebrew, M.A. et al. Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 36, 276–287 (2012).
Bennett, C.L. & Clausen, B.E. DC ablation in mice: promises, pitfalls, and challenges. Trends Immunol. 28, 525–531 (2007).
Meredith, M.M. et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209, 1153–1165 (2012).
Satpathy, A.T. et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209, 1135–1152 (2012).
Hildner, K. et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008).
Lewis, K.L. et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity 35, 780–791 (2011).
Swiecki, M. et al. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8+ T cell accrual. Immunity 33, 955–966 (2010).
Hashimoto, D., Miller, J. & Merad, M. Dendritic cell and macrophage heterogeneity in vivo. Immunity 35, 323–335 (2011).
Torti, N. et al. Batf3 transcription factor-dependent DC subsets in murine CMV infection: differential impact on T-cell priming and memory inflation. Eur. J. Immunol. 41, 2612–2618 (2011).
Mashayekhi, M. et al. CD8a+ dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity 35, 249–259 (2011).
Cervantes-Barragan, L. et al. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc. Natl. Acad. Sci. USA 109, 3012–3017 (2012).
Bogunovic, M. et al. Origin of the lamina propria dendritic cell network. Immunity 31, 513–525 (2009).
Varol, C. et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity 31, 502–512 (2009).
Ohl, L. et al. CCR7 governs skin dendritic cell migration under inflammatory and steady-state conditions. Immunity 21, 279–288 (2004).
Jakubzick, C. et al. Lymph-migrating, tissue-derived dendritic cells are minor constituents within steady-state lymph nodes. J. Exp. Med. 205, 2839–2850 (2008).
Randolph, G.J., Ochando, J. & Partida-Sanchez, S. Migration of dendritic cell subsets and their precursors. Annu. Rev. Immunol. 26, 293–316 (2008).
McKenna, H.J. et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood 95, 3489–3497 (2000).
Boring, L. et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C–C chemokine receptor 2 knockout mice. J. Clin. Invest. 100, 2552–2561 (1997).
Zigmond, E. et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37, 1076–1090 (2012).
Dudziak, D. et al. Differential antigen processing by dendritic cell subsets in vivo. Science 315, 107–111 (2007).
Radtke, F., Fasnacht, N. & MacDonald, H.R. Notch signaling in the immune system. Immunity 32, 14–27 (2010).
Edelson, B.T. et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207, 823–836 (2010).
McDonald, K.G. et al. Dendritic cells produce CXCL13 and participate in the development of murine small intestine lymphoid tissues. Am. J. Pathol. 176, 2367–2377 (2010).
Caton, M.L., Smith-Raska, M.R. & Reizis, B. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204, 1653–1664 (2007).
Waskow, C. et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 9, 676–683 (2008).
Kabashima, K. et al. Intrinsic lymphotoxin-β receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity 22, 439–450 (2005).
Summers deLuca, L. & Gommerman, J.L. Fine-tuning of dendritic cell biology by the TNF superfamily. Nat. Rev. Immunol. 12, 339–351 (2012).
Fütterer, A. et al. The lymphotoxin β receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity 9, 59–70 (1998).
Tussiwand, R. et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490, 502–507 (2012).
Bajaña, S. et al. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol. 189, 3368–3377 (2012).
Klein, U. et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7, 773–782 (2006).
Manieri, N.A. et al. Igf2bp1 is required for full induction of Ptgs2 mRNA in colonic mesenchymal stem cells in mice. Gastroenterology 143, 110–121 (2012).
Brown, S.L. et al. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J. Clin. Invest. 117, 258–269 (2007).
Satpathy, A.T. et al. Re(de)fining the dendritic cell lineage. Nat. Immunol. 13, 1145–1154 (2012).
Possot, C. et al. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat. Immunol. 12, 949–958 (2011).
Lee, J.S. et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13, 144–151 (2012).
Ota, N. et al. IL-22 bridges the lymphotoxin pathway with the maintenance of colonic lymphoid structures during infection with Citrobacter rodentium. Nat. Immunol. 12, 941–948 (2011).
Wang, Y. et al. Lymphotoxin beta receptor signaling in intestinal epithelial cells orchestrates innate immune responses against mucosal bacterial infection. Immunity 32, 403–413 (2010).
Kim, Y.G. et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity 34, 769–780 (2011).
Rivollier, A. et al. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J. Exp. Med. 209, 139–155 (2012).
Basu, R. et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity 37, 1061–1075 (2012).
Steinman, R.M. Decisions about dendritic cells: past, present, and future. Annu. Rev. Immunol. 30, 1–22 (2011).
Heng, T.S. & Painter, M.W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002).
Yu, H. et al. APP processing and synaptic plasticity in presenilin-1 conditional knockout mice. Neuron 31, 713–726 (2001).
Yin, L. et al. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science 291, 2162–2165 (2001).
Keskintepe, L. et al. Derivation and comparison of C57BL/6 embryonic stem cells to a widely used 129 embryonic stem cell line. Transgenic Res. 16, 751–758 (2007).
Schwenk, F., Baron, U. & Rajewsky, K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080–5081 (1995).
Robben, P.M. et al. Production of IL-12 by macrophages infected with Toxoplasma gondii depends on the parasite genotype. J. Immunol. 172, 3686–3694 (2004).
Akashi, K. et al. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000).
Onai, N. et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8, 1207–1216 (2007).
Acknowledgements
We thank B. Sleckman (Washington University in St. Louis) for Nik−/− mice; T. Watts (University of Toronto) for Ifnar1−/− mice; J. Boothroyd (Stanford University) for the plasmid PRU-FLuc-GFP; the Immunological Genome Project consortium for use of their database54; and the Alvin J. Siteman Cancer Center at Washington University School of Medicine for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility. Supported by the Howard Hughes Medical Institute, the US National Institutes of Health (AI076427-02 to K.M.M., R01 GM55479 to R.K., R01 DE021255-01 and U01 AI095542-01 to M.C., R01 DK071619 to T.S.S. and R01 DK064798 to R.D.N.), the US Department of Defense (W81XWH-09-1-0185 to K.M.M.), the American Heart Association (12PRE8610005 to A.T.S. and 12PRE12050419 to W.K.), the Canadian Institutes of Health Research (MOP 67157 to J.L.G. and FRN 11530 to C.J.G.) and the National Cancer Institute (P30 CA91842 for the Alvin J. Siteman Cancer Center).
Author information
Authors and Affiliations
Contributions
A.T.S. and K.M.M. designed the study; A.T.S., C.G.B., J.S.L. and C.S. did experiments related to infection with C. rodentium, with guidance from W.O., M.C. and K.M.M.; A.T.S., C.G.B. and D.N. did experiments related to Ltbr−/− mice, with guidance from C.J.G. and J.L.G.; N.A.M. did experiments related to wound healing, with guidance from T.S.S.; A.T.S. and X.W. did microarray analysis; A.T.S., S.R.T., W.K., W.-L.L., M.T., T.L.M. and K.G.M. did experiments related to cDC development in mice deficient in Notch2, Irf4 or Batf3, with guidance from R.K., R.D.N. and K.M.M.; A.T.S., C.G.B. and M.M.M. did experiments with Zbtb46gfp and Zbtb46DTR mice, with guidance from M.C.N. and K.M.M.; and A.T.S. and K.M.M. wrote the manuscript with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
W.O. is an employee of Genentech.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Table 1 (PDF 18525 kb)
Rights and permissions
About this article
Cite this article
Satpathy, A., Briseño, C., Lee, J. et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol 14, 937–948 (2013). https://doi.org/10.1038/ni.2679
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2679
This article is cited by
-
Distal colonocytes targeted by C. rodentium recruit T-cell help for barrier defence
Nature (2024)
-
Signaling pathways involved in the biological functions of dendritic cells and their implications for disease treatment
Molecular Biomedicine (2023)
-
Testicular macrophages are recruited during a narrow fetal time window and promote organ-specific developmental functions
Nature Communications (2023)
-
Transitional dendritic cells are distinct from conventional DC2 precursors and mediate proinflammatory antiviral responses
Nature Immunology (2023)
-
The transcription factor Zeb1 controls homeostasis and function of type 1 conventional dendritic cells
Nature Communications (2023)