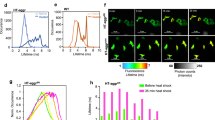
Abstract
The extensive links between proteotoxic stress, protein aggregation and pathologies ranging from ageing to neurodegeneration underscore the importance of understanding how cells manage protein misfolding. Using live-cell imaging, we determine the fate of stress-induced misfolded proteins from their initial appearance until their elimination. Upon denaturation, misfolded proteins are sequestered from the bulk cytoplasm into dynamic endoplasmic reticulum (ER)-associated puncta that move and coalesce into larger structures in an energy-dependent but cytoskeleton-independent manner. These puncta, which we name Q-bodies, concentrate different misfolded and stress-denatured proteins en route to degradation, but do not contain amyloid aggregates, which localize instead to the insoluble protein deposit compartment. Q-body formation and clearance depends on an intact cortical ER and a complex chaperone network that is affected by rapamycin and impaired during chronological ageing. Importantly, Q-body formation enhances cellular fitness during stress. We conclude that spatial sequestration of misfolded proteins in Q-bodies is an early quality control strategy occurring synchronously with degradation to clear the cytoplasm of potentially toxic species.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008).
Lindquist, S. L. & Kelly, J. W. Chemical and biological approaches for adapting proteostasis to ameliorate protein misfolding and aggregation diseases: progress and prognosis. Cold Spring Harb. Perspect. Biol. 3, 1–34 (2011).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid, and human disease. Ann. Rev. Biochem. 75, 333–366 (2006).
Houck, S.A., Singh, S. & Cyr, D. M. Cellular responses to misfolded proteins and protein aggregates. Methods Mol. Biol. 832, 455–461 (2012).
Chen, B., Retzlaff, M., Roos, T. & Frydman, J. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 3, a004374 (2011).
Kaganovich, D., Kopito, R. & Frydman, J. Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 (2008).
Specht, S., Miller, S. B., Mogk, A. & Bukau, B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J. Cell Biol. 195, 617–629 (2011).
Malinovska, L., Kroschwald, S., Munder, M. C., Richter, D. & Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 23, 3041–3056 (2012).
Johnston, J. A., Ward, C. L. & Kopito, R. R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898 (1998).
Zhang, X. & Qian, S. B. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol. Biol. Cell 22, 3277–3288 (2011).
Douglas, P. M., Summers, D. W. & Cyr, D. M. Molecular chaperones antagonize proteotoxicity by differentially modulating protein aggregation pathways. Prion 3, 51–58 (2009).
Cohen, E., Bieschke, J., Perciavalle, R. M., Kelly, J. W. & Dillin, A. Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 (2006).
Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2004).
Liu, B. et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 140, 257–267 (2010).
Nakatogawa, H., Ichimura, Y. & Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 (2007).
Sheth, U. & Parker, R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125, 1095–1109 (2006).
Toshima, J. Y. et al. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc. Natl Acad. Sci. USA 103, 5793–5798 (2006).
Wright, R., Basson, M., D’Ari, L. & Rine, J. Increased amounts of HMG-CoA reductase induce ”karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 107, 101–114 (1988).
Shibata, Y., Voeltz, G. K. & Rapoport, T. A. Rough sheets and smooth tubules. Cell 126, 435–439 (2006).
Voeltz, G. K., Prinz, W. A., Shibata, Y., Rist, J. M. & Rapoport, T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586 (2006).
Shibata, Y. et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 283, 18892–18904 (2008).
West, M., Zurek, N., Hoenger, A. & Voeltz, G. K. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193, 333–346 (2011).
McDonough, H. & Patterson, C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 8, 303–308 (2003).
Mayer, M. P. & Bukau, B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684 (2005).
McClellan, A. J., Scott, M. D. & Frydman, J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121, 739–748 (2005).
Coppinger, J. A. et al. A chaperone trap contributes to the onset of cystic fibrosis. PLoS One 7, e37682 (2012).
Schneider, C. et al. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc. Natl Acad. Sci. USA 93, 14536–14541 (1996).
Becker, J., Walter, W., Yan, W. & Craig, E. A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell Biol. 16, 4378–4386 (1996).
Caplan, A. J., Tsai, J., Casey, P. J. & Douglas, M. G. Farnesylation of YDJ1p is required for function at elevated growth temperatures in Saccharomyces cerevisiae. J. Biol. Chem. 267, 18890–18895 (1992).
Flom, G. A., Lemieszek, M., Fortunato, E. A. & Johnson, J. L. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol. Biol. Cell 19, 5249–5258 (2008).
Kampinga, H. H. & Craig, E. A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 (2010).
Youker, R. T., Walsh, P., Beilharz, T., Lithgow, T. & Brodsky, J. L. Distinct rolesfor the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 15, 4787–4797 (2004).
Shorter, J. & Lindquist, S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 27, 2712–2724 (2008).
Tipton, K. A., Verges, K. J. & Weissman, J. S. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell 32, 584–591 (2008).
Gupta, R. et al. Firefly luciferase mutants as sensors of proteome stress. Nat. Methods 8, 879–884 (2011).
Chernoff, Y. O., Lindquist, S. L., Ono, B., Inge-Vechtomov, S. G. & Liebman, S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi +]. Science 268, 880–884 (1995).
Sondheimer, N. & Lindquist, S. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell 5, 163–172 (2000).
Meriin, A. B. et al. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 157, 997–1004 (2002).
Piper, P. W. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 134, 121–127 (1995).
Trotter, E. W. et al. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 277, 44817–44825 (2002).
Morimoto, R. I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438 (2008).
Conn, C. S. & Qian, S. B. mTOR signaling in protein homeostasis: less is more? Cell Cycle 10, 1940–1947 (2011).
Taylor, R. C. & Dillin, A. Aging as an event of proteostasis collapse. Cold Spring Harb. Perspect. Biol. 3, 1–17 (2011).
Fabrizio, P. & Longo, V. D. The chronological life span of Saccharomyces cerevisiae. Aging Cell 2, 73–81 (2003).
Narayanaswamy, R. et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl Acad. Sci. USA 106, 10147–10152 (2009).
Peters, T. W. et al. Tor1 regulates protein solubility in Saccharomyces cerevisiae. Mol. Biol. Cell 23, 4679–4688 (2012).
Kapahi, P. et al. With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453–465 (2010).
Glover, J. R. & Lindquist, S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998).
Mandal, A. K. et al. Hsp110 chaperones control client fate determination in the hsp70-Hsp90 chaperone system. Mol. Biol. Cell 21, 1439–1448 (2010).
Huyer, G. et al. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 279, 38369–38378 (2004).
Ouellet, J. & Barral, Y. Organelle segregation during mitosis: lessons from asymmetrically dividing cells. J. Cell Biol. 196, 305–313 (2012).
Zhou, C. et al. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell 147, 1186–1196 (2011).
Gidalevitz, T., Krupinski, T., Garcia, S. & Morimoto, R. I. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. 5, e1000399 (2009).
Olzscha, H. et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144, 67–78 (2011).
Kikis, E. A., Gidalevitz, T. & Morimoto, R. I. Protein homeostasis in models of aging and age-related conformational disease. Adv. Exp. Med. Biol. 694, 138–159 (2010).
Shorter, J. Hsp104: a weapon to combat diverse neurodegenerative disorders. Neurosignals 16, 63–74 (2008).
Gidalevitz, T., Prahlad, V. & Morimoto, R. I. The stress of protein misfolding: from single cells to multicellular organisms. Cold Spring Harb. Perspect. Biol. 3, 1–18 (2011).
Durieux, J., Wolff, S. & Dillin, A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91 (2011).
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Nathan, D. F. & Lindquist, S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell Biol. 15, 3917–3925 (1995).
Kaksonen, M., Toret, C. P. & Drubin, D. G. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123, 305–320 (2005).
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Alberti, S., Gitler, A. D. & Lindquist, S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919 (2007).
Furuta, N., Fujimura-Kamada, K., Saito, K., Yamamoto, T. & Tanaka, K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell 18, 295–312 (2007).
Acknowledgements
We thank C. Toret and V. Albanese for experimental advice and discussions; and C. Toret and R. Andino for critical reading of the manuscript. S.E-T. was initially supported by a fellowship from Fondation pour la Recherche Medicale (France). W.I.M.V. was supported by the Marie Curie International Outgoing Fellowship Programme. This work was supported by grants from the NIH and a Senior Scholar Award from the Ellison Foundation to J.F.
Author information
Authors and Affiliations
Contributions
S.E-T. and J.F. conceived the project, S.E-T. performed most experiments, W.I.M.V. performed experiments in Fig. 1d,e, Fig. 3e,f and Supplementary Fig. S1b, and all authors interpreted the experiments and contributed to writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Formation of Q-bodies.
(a) WT cells expressing Ubc9ts–GFP were grown at 28 °C in galactose medium and shifted at 33 °C or 37 °C in glucose medium. 5 min series images shows Ubc9ts–GFP signal over a 30 min movie. Scale bars equal 1 μm. (b) pdr5Δ and pdr5Δatg8Δ cells expressing Ubc9ts–GFP were grown as in a. Upon temperature shift, cells were treated with 100 μM MG132 as indicated. Ubc9ts–GFP was analyzed by immunoblot using anti-GFP antibodies. (c) Images of Ubc9-GFP expressed in WT cells at 28 °C (left panel) and shifted for 15 min at 37 °C. GFP (right panel). Scale bars equal 1.5 μm.
Supplementary Figure 2 Q-bodies associate with specific sub-cellular structures.
(a) Two-color images of Z-focal plans (0.2 μm intervals; obtained after 10 min at 37 °C) of cells expressing Ubc9ts–CHFP (red) or Ubc9ts–GFP (green) and the following markers: GFP–Snc1 (early endosomes; green), GFP–Pep12 (late endosomes; green), CHFP–Atg8 (autophagic vesicles; red); the vacuole was imaged by treating cells with MDY64 (blue). Scale bars equal 1 μm. (b) Cells expressing Spc42–GFP (green) and Ubc9ts–CHFP (red) were imaged 15 min after a shift from 28 °C to 37 °C. Scale bars equal 1 μm.
Supplementary Figure 3 Hsp70–Hsp90 chaperones promote the maturation and degradation of Q-bodies.
(a) Schematic of the Hsp70 chaperone system and its connection to the Hsp90 system. (b) Ubc9ts–GFP was expressed in WT and ssa1-4 ssa2Δssa3Δssa4Δ (herein ssa1ts ssa2Δssa3Δssa4Δ) background at 28 °C in galactose medium and shifted at 37 °C in glucose medium. 5 min series of images shows Ubc9ts–GFP in these strains over 30 min. Scale bars equal 1 μm. (c) Representation of the average number of puncta per cell in the WT (purple squares) and the ydj1Δ (orange circles) strains over time. Puncta assessed from a total population of n = 38 cells over three independent experiments (1 field counted per experiment). (*) p<0.05 (**) p<0.005 compared to WT for the same indicated time. (d) Ubc9ts–GFP expressed in WT, hlj1Δ and hlj1Δydj1-151 cells was imaged as in b. Scale bar equals 1 μm. (e) Average number of puncta per cell in the WT (purple squares) and the sse1Δ (green triangles) strains over timePuncta assessed from a total population of n = 16 cells over three independent experiments (1 field counted per experiment). (**) p<0.005 compared to the WT for the same indicated time.
Supplementary Figure 4 Hsp104 does not colocalize with perinuclear Q-bodies.
Images of a fixed cell expressing Hsp104–GFP (green) and Ubc9ts–CHFP (red) and stained with DAPI (blue) after a 15 min shift from 28 °C to 37 °C. Merged represents the overlay of the three channels. Scale bar equals 1 μm.
Supplementary Figure 5 Different types of misfolded proteins, but not amyloids, co-localize to and are processed together via the same Q-bodies.
(a) WT cells expressing Ubc9ts–CHFP and Luc-GFP were grown at 28 °C in galactose medium and imaged at 37 °C in glucose medium. 5 min series of images show Ubc9ts–CHFP in red and Luc-GFP in green. Cells expressing Ubc9-GFP were similarly prepared and imaged 15 min after the shift. Scale bar equals 1 μm. (b) WT cells expressing CHFP-VHL were grown as in a. 5 min series of images shows CHFP-VHL over 30 min. Scale bar equals 1 μm. (c) WT cells expressing GFP-VHL (green) and Rnq1-CHFP (red) were grown and imaged as in a. Scale bar equals 1 μm. (d) Cells expressing Hsp42–GFP and Htt-Q97-CHFP were grown at 28 °C in galactose medium and shifted for 10 min at 37 °C in glucose medium. Two-color images (deconvolved in the lower panel) show Hsp42–GFP signal in green and Htt-Q97-CHFP signal in red. Scale bar equals 1 μm. (e) Cells expressing Hsp104–GFP (green) and Htt-Q97-CHFP (red) were grown and imaged as in d. Scale bars equal 1 μm.
Supplementary Figure 6 Aging impairs the Q-body pathway.
Cells expressing Ubc9ts–GFP were grown at 28 °C for 5 hours (young) or for 7 days (aged) and imaged at 37 °C. 5 min series of a 30 min movie show the GFP signal. Scale bars represent 1 μm.
Supplementary Figure 7 Deletion of Hsp42, Hsp26 or Hsp104 does not cause any appreciable growth defects at 37 °C.
A suspension of WT, hsp104Δ, hsp42Δ, and hsp26Δ cells were serially diluted and dropped on YPD at 28 °C and 37 °C.
Supplementary Figure 8 Full scans of original immunoblot data presented in this study.
Red outlines represent the immunoblot sections presented in the corresponding main figures.
Supplementary information
Supplementary Information
Supplementary Information (PDF 689 kb)
Supplementary Table 1
Supplementary Information (XLSX 15 kb)
Misfolded Ubc9ts forms Q-bodies.
Live-cell movie of Ubc9ts–GFP in a WT cell at 37 °C. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 1f. (AVI 2678 kb)
Actin cytoskeleton does not affect the Q-body pathway.
Live-cell movie of cells expressing Ubc9ts–GFP (left panel) or Abp1–GFP (right panel) at 37 °C with (lower panel) or without (upper panel) LatA treatment. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 2a. (AVI 13424 kb)
The Q-body pathway is energy dependent.
Live-cell movie of Ubc9ts–GFP in WT cell at 37 °C with (right panel) or without (left panel) sodium azide and deoxyglucose (Az/Deox) treatment. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 2d. (AVI 6903 kb)
Q-bodies localize in proximity to the cortical ER.
3D projection of a live-cell expressing Ubc9ts–CHFP Q-bodies (red) and Rtn1-GFP (green, cortical ER) at 37 °C. 0.2 μm intervals. Refers to Fig. 3c. (AVI 2388 kb)
The Q-body pathway depends on an intact cortical ER.
Live-cell movie of Ubc9ts–GFP in WT and rtn1Δrtn2Δyop1Δ cells at 37 °C. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 3e. (AVI 5395 kb)
Hsp70 is required for Q-body formation, dynamics and clearance.
Live-cell movie of Ubc9ts–GFP in WT, ssa1Δssa2Δ and sse1Δ cells at 37 °C. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 4c and h. (AVI 9091 kb)
Hsp82 is required for Q-body maturation and clearance.
Live-cell movie of Ubc9ts–GFP in HSP82 and hsp82ts cells at 37 °C. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 4e. (AVI 6113 kb)
Ydj1 participates in the formation and localization of Q-bodies.
Live-cell movie of Ubc9ts–GFP expressed in ydj1Δ cells with an empty vector, YDJ1, or ydj1(C406S) at 37 °C. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 4g. (AVI 14555 kb)
Hsp104 co-localizes with peripheral Ubc9ts-forming Q-bodies.
Live-cell movie of a cell co-expressing Ubc9ts–CHFP (red) and Hsp104–GFP (green) at 37 °C. Interval between frames is 15 s. Total time of acquisition was 60 min. Refers to Fig. 5a. (AVI 19157 kb)
Q-body pathway relies on the balance between Hsp104 and Hsp42 activities.
Live-cell movie of Ubc9ts–GFP in WT, hsp104Δ, hsp42Δ, and hsp42Δhsp104Δ cells at 37 °C. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 5d and g. (AVI 13544 kb)
VHL, but not Htt-Q97, is concentrated to Q-bodies.
Live-cell movie of Ubc9ts–GFP (green) co-expressed with CHFP-VHL (red) or Htt-Q97-CHFP (red) in WT cell at 37 °C. Interval between frames is 15 s. Total time of acquisition was 30 min. Refers to Fig. 6a and b. (AVI 16100 kb)
TOR signaling regulates the Q-body pathway.
Live-cell movie of Ubc9ts–GFP in WT cell with or without 0.2 μg ml−1 rapamycin (Rap) treatment at 37 °C. Total time of acquisition was 30 min. Refers to Fig. 7c. (AVI 5395 kb)
Rights and permissions
About this article
Cite this article
Escusa-Toret, S., Vonk, W. & Frydman, J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol 15, 1231–1243 (2013). https://doi.org/10.1038/ncb2838
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2838
This article is cited by
-
Nuclear and cytoplasmic spatial protein quality control is coordinated by nuclear–vacuolar junctions and perinuclear ESCRT
Nature Cell Biology (2023)
-
Loss of PML nuclear bodies in familial amyotrophic lateral sclerosis-frontotemporal dementia
Cell Death Discovery (2023)
-
Artificial Hsp104-mediated systems for re-localizing protein aggregates
Nature Communications (2023)
-
Metamorphism in TDP-43 prion-like domain determines chaperone recognition
Nature Communications (2023)
-
The GET pathway is a major bottleneck for maintaining proteostasis in Saccharomyces cerevisiae
Scientific Reports (2023)