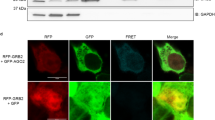
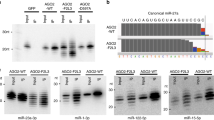
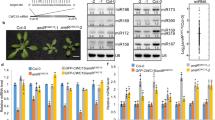
Abstract
MicroRNAs (miRNAs) form a class of short RNAs (∼ 21 nucleotides) that post-transcriptionally regulate partially complementary messenger RNAs. Each miRNA may target tens to hundreds of transcripts to control key biological processes. Although the biochemical reactions underpinning miRNA biogenesis and activity are relatively well defined1,2 and the importance of their homeostasis is increasingly evident, the processes underlying regulation of the miRNA pathway in vivo are still largely elusive3. Autophagy, a degradative process in which cytoplasmic material is targeted into double-membrane vacuoles, is recognized to critically contribute to cellular homeostasis. Here, we show that the miRNA-processing enzyme, DICER (also known as DICER1), and the main miRNA effector, AGO2 (also known as eukaryotic translation initiation factor 2C, 2 (EIF2C2)), are targeted for degradation as miRNA-free entities by the selective autophagy receptor NDP52 (also known as calcium binding and coiled-coil domain 2 (CALCOCO2)). Autophagy establishes a checkpoint required for continued loading of miRNA into AGO2; accordingly, NDP52 and autophagy are required for homeostasis and activity of the tested miRNAs. Autophagy also engages post-transcriptional regulation of the DICER mRNA, underscoring the importance of fine-tuned regulation of the miRNA pathway. These findings have implications for human diseases linked to misregulated autophagy, DICER- and miRNA-levels, including cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
10 July 2015
In the version of this Letter originally published, the TUBA immunoblotting panel and the Coomassie-stained panel was used in Figure 5c (in the dataset corresponding to let-7 antagomir treatment, top set of panels), and reused in Figure 5e, without appropriate acknowledgement. The Coomassie-stained gel was vertically flipped in Figure 5e but the alignment of the lanes was maintained. The TUBA immunoblot and Coomassie-stained membranes represent experimental controls. The p62 panel in Figures 2j and 2k (CQ-treated) was also reused without appropriate attribution. In all cases of reuse of blots between panels, the samples were obtained within one representative experiment and processed in parallel. The authors confirm that all instances of vertical splicing of lanes, for example in Figs 1 and 3, were carried out in full compliance with the journal guidelines. All spliced samples were collected and processed in a single experiment. The original publication was missing Supplementary Fig. S3 containing the uncropped scans of the blots; this has now been included online.
References
Kim, V. N., Han, J. & Siomi, M. C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–39 (2009).
Krol, J., Loedige, I. & Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Gene. 11, 597–610 (2010).
Siomi, H. & Siomi, M. C. Post transcriptional regulation of microRNA biogenesis in animals. Mol. Cell 38, 323–32 (2010).
Eulalio, A., Huntzinger, E. & Izaurralde, E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 15, 346–53 (2008).
Bazzini, A. A., Lee, M. T. & Giraldez, A. J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237 (2012).
Ameres, S. L. et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–9 (2010).
Chatterjee, S. & Grosshans, H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461, 546–9 (2009).
Johnston, M., Geoffroy, M. C., Sobala, A., Hay, R. & Hutvagner, G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol. Biol. Cell 21, 1462–9 (2010).
Mayr, C. & Bartel, D. P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–84 (2009).
Vaucheret, H., Vazquez, F., Crete, P. & Bartel, D. P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–97 (2004).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–41 (2011).
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–45 (2007).
Thurston, T. L., Wandel, M. P., von Muhlinen, N., Foeglein, A. & Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–8 (2012).
Shaid, S., Brandts, C. H., Serve, H. & Dikic, I. Ubiquitination and selective autophagy. Cell Death Diff.http://dx.doi.org/10.1038 (2012).
Watanabe, Y. & Tanaka, M. p62/SQSTM1 in autophagic clearance of a non-ubiquitylated substrate. J. Cell Sci. 124, 2692–701 (2011).
Gibbings, D. J., Ciaudo, C., Erhardt, M. & Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11, 1143–9 (2009).
Haase, A. D. et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6, 961–7 (2005).
Lee, Y. S. et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 11, 1150–6 (2009).
Gibbings, D. & Voinnet, O. Control of RNA silencing and localization by endolysosomes. Trends Cell Biol. 20, 491–501 (2010).
Friend, K. et al. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat. Struct. Mol. Biol. 19, 176–83 (2012).
Pare, J. M. et al. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol. Biol. Cell 20, 3273–84 (2009).
Liu, J., Valencia-Sanchez, M. A., Hannon, G. J. & Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7, 719–23 (2005).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–26 (2010).
Marzella, L., Ahlberg, J. & Glaumann, H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J. Cell Biol. 93, 144–54 (1982).
Di, Y. et al. HCC-associated protein HCAP1, a variant of GEMIN4, interacts with zinc-finger proteins. J. Biochem. 133, 713–8 (2003).
Meister, G. et al. Identification of novel argonaute-associated proteins. Curr. Biol. 15, 2149–55 (2005).
Mourelatos, Z. et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16, 720–8 (2002).
Gantier, M. P. et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 39, 5692–703 (2011).
Chendrimada, T. P. et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–4 (2005).
Sakurai, K. et al. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 39, 1510–25 (2011).
Noland, C. L., Ma, E. & Doudna, J. A. siRNA repositioning for guide strand selection by human Dicer complexes. Mol. Cell 43, 110–21 (2011).
Iwasaki, S. et al. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 39, 292–9 (2010).
Mamane, Y., Petroulakis, E., LeBacquer, O. & Sonenberg, N. mTOR, translation initiation and cancer. Oncogene 25, 6416–22 (2006).
Doench, J. G. & Sharp, P. A. Specificity of microRNA target selection in translational repression. Genes Dev. 18, 504–11 (2004).
Slack, F. let-7 microRNA reduces tumor growth. Cell Cycle 8, 1823 (2009).
Obad, S. et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 43, 371–8 (2011).
Csorba, T., Lozsa, R., Hutvagner, G. & Burgyan, J. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. Cell Mol. Biol. 62, 463–72 (2010).
Derrien, B. et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl Acad. Sci. USA 109, 15942–15946 (2012).
Wander, S. A., Hennessy, B. T. & Slingerland, J. M. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J. Clin. Inv. 121, 1231–41 (2011).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–8 (2005).
Rudel, S., Flatley, A., Weinmann, L., Kremmer, E. & Meister, G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA 14, 1244–53 (2008).
Eystathioy, T. et al. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13, 1338–51 (2002).
Doench, J. G., Petersen, C. P. & Sharp, P. A. siRNAs can function as miRNAs. Gen. Dev. 17, 438–42 (2003).
Klionsky, D. J., Elazar, Z., Seglen, P. O. & Rubinsztein, D. C. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4, 849–950 (2008).
Jakymiw, A. et al. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 7, 1267–74 (2005).
Mesquita, F. S. et al. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 8, e1002743 (2012).
Costes, S. V. et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86, 3993–4003 (2004).
Spiegelhalter, C. et al. From dynamic live cell imaging to 3D ultrastructure: novel integrated methods for high pressure freezing and correlative light-electron microscopy. PLoS ONE 5, e9014 (2010).
Sahu, R. et al. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 20, 131–9 (2011).
Chi, S. W., Zang, J. B., Mele, A. & Darnell, R. B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–86 (2009).
Acknowledgements
The authors would like to thank M. Johnston (ETH-Zurich, Switzerland) for providing HEK293T cells stably transfected with Tet-inducible Flag–AGO2 as well as the protocol for detection of ubiquitylated Flag–AGO2. Financial support was provided by a core grant from ETH-Z to O.V., and the Pasteur Institute to P.C. S.M. is a Wellcome Trust Research Career Development Fellow. The authors thank K. McGourty and D. Li for helpful discussions.
Author information
Authors and Affiliations
Contributions
D.G. conceived the hypothesis. D.G., S.M., F.J. and Y.S. performed and analysed experiments. D.G., S.M. and O.V. designed the overall research. D.G., S.M., P.C. and O.V. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 362 kb)
Rights and permissions
About this article
Cite this article
Gibbings, D., Mostowy, S., Jay, F. et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol 14, 1314–1321 (2012). https://doi.org/10.1038/ncb2611
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2611
This article is cited by
-
The helicase domain of human Dicer prevents RNAi-independent activation of antiviral and inflammatory pathways
The EMBO Journal (2024)
-
microRNAs in action: biogenesis, function and regulation
Nature Reviews Genetics (2023)
-
AMPK-autophagy-mediated inhibition of microRNA-30a-5p alleviates morphine tolerance via SOCS3-dependent neuroinflammation suppression
Journal of Neuroinflammation (2022)
-
Autophagy regulation by RNA alternative splicing and implications in human diseases
Nature Communications (2022)
-
SQSTM1/p62 promotes miR-198 loading into extracellular vesicles and its autophagy-related secretion
Human Cell (2022)